Single nucleotide polymorphisms of MAGE-A3 gene and its clinical implications in Chinese patients with non-small cell lung cancer (NSCLC)
Introduction
MAGE-A3 gene is a member of the cancer/testis (CT) gene families. The locus for the MAGE-A3 gene is at Xq28. MAGE-A3 gene has three exons and high homology with the other members of MAGE-A gene family (1). The protein it encodes is a cytoplasmic protein which had a molecular weight with 48,000 and composed of 314 amino acids (2). MAGE-A3 is a tumor common antigen and widely expressed in tumor cells of various tissue types, but not expressed in the normal tissue cells (except testis and placenta). MAGE-A3 protein was considered to be a true tumor specific antigen and found expressed in many malignancies such as melanoma, non-small cell lung cancer (NSCLC), bladder cancer, head and neck neoplasm, esophageal squamous cell carcinoma and hepatocellular carcinoma and so on (3). It has been reported that the expression of MAGE-A3 antigen in NSCLC was over 50% (4,5). Studies has shown that MAGE-A3 gene expression correlated to tumor TNM staging and clinical prognosis of tumor, the more advanced tumor tissue has a higher expression of MAGE-A3, and MAGE-A3 gene expression predicted a poor clinical prognosis for patients with NSCLC (6,7). MAGE-A3 has a lots of gene polymorphisms, but there was no data concerning whether it could be used for predicting the survival of NSCLC patients as the tumor prognosis factor. To confirm the role of MAGE-A3 gene polymorphisms in prognosis for patients, we analyzed the allele frequency of genotypes on single nucleotide polymorphism (SNP) loci with MAGE-A3 gene and the relations between gene polymorphisms and clinical parameters, such as gender, age, smoking status and pathological classification.
Materials and methods
Patients
A total of 191 patients with pathologically confirmed NSCLC, which were reviewed from January, 2003 to December, 2010 at Guangdong General Hospital (GGH). Their corresponding clinical information was from the electronic medical record database of Guangdong Lung Cancer Institute (GLCI). Patients with pathologically adenocarcinoma occupied 50.3% (96/191) and squamous carcinoma occupied 49.7% (95/191). The average age is 60.91 years, ranging from 26 to 82. 71.2% (136/191) of patients was men and other 28.8% (55/191) were women. Tumor specimens were retrieved from the GLCI tumor tissue bank. The study was approved by the institutional review boards of GGH. All patients provided informed consent. The last follow up was September 4th, 2012.
DNA/RNA isolation, reverse transcription, real-time qRT-PCR, and sequencing
Total DNA /RNA was isolated from different tumors using QIAampDNA Mini Kit and RNeasy Mini Kit (QIAGEN).
cDNA for qRT-PCR was synthesized using the Superscript II RNase H reverse transcriptase kit (Invitrogen).
The mRNA expression level of epidermal growth factor receptor (EGFR) and the β2M reference gene was determined by Taqman probe real-time PCR. PCR was performed as previously described (7). The sequences for the primers used in qRT-PCR were as follows: F1: 5'- CTGGAACGGTGAAGGTGACA -3' and R1: 5'-CGGCCACATTGTGAACTTTG-3' and Probe-1: 5'-Vic-TGCTCGCTCC AACC-MGB-3' for β2M gene amplification and F2: 5'-GGCTCAGATAGTGCC AACGGTG-3' and R2: 5'-CTTCCATCAGCTCGATGCCAAA-3' for genotypes detection on five SNP loci and F3: 5’-CAAATGAGCTGGCAAGTGCCGTGTCCTG-3' and R3: 5'-GAGTTTCCCAAACACTCAGTGAAACA-3' for EGFR exon 18 point mutation detection and F4: 5'-GCAATATCAGCCTTAGGTGCGGC-3’ and R4: 5'-CATAGAAAGTG AACATTTAGGATGT-3' for exon 19 deletion detection and F5: 5'-CCATGAGTACGTATTTTGAAACTCA-3' and R5: 5'-CATATCCCCATG GCAAA CTCTT-3' for exon 20 point mutation detection and F6: 5'-ATGAACATGA CCCTGAATTCGG-3' and R6:5'-GCTCACCCAGAATGTCTGGA-3' for exon 21 point mutation detection.
PCR product was recovered by agarose gel DNA purification kit (Takara) and sent for sequencing analysis using the 3,700 DNA Genetic Analyzer (Applied Biosystems). The resulting data were analyzed using the BioEdit Analysis software and validated by comparing with the NCBI gene bank.
Statistical analysis
Chi-square or continuity correction tests were used to compare the differences in SNP loci genotypes of MAGE-A3 gene and EGFR expression and clinical parameters. Overall survival (OS) was calculated from commencement of pathologically confirmed NSCLC to the last visit or death from any cause. Survival curves among patient groups were compared by the Log-rank test. All statistical tests were two sided and P<0.05 was considered to be statistically significant.
Results
Sequencing results of MAGE-A3 polymorphisms and EGFR mutations
Sequencing results showed that, in all 191 patients, there have five locus existed SNP polymorphisms, just except one patient haven’t detected any genotypes. Five locus including loci rs5970360, rs5925210, rs5970361, rs5925211 and rs35123853. Genotypes of the five SNP locus were CC/CT, CC/CG, CC/CA, AT/AA and GG/GA (Figure 1A-C). Allele frequencies were 0.681 (CC, 130/191) and 0.319 (CT, 61/191) for the SNP locus rs5970360, and 0.660 (CC, 126/191) and 0.340 (CG, 65/191) for the SNP locus rs5925210, and 0.686 (CC, 131/191) and 0.314 (CA, 60/191) for the SNP locus rs5970361, and 0.016 (AT, 3/191) and 0.984 (AA, 188/191) for the SNP locus rs5925211, and 1.000 (GG, 191/191) and 0.000 (GA, 0/191) for the SNP locus rs35123853.
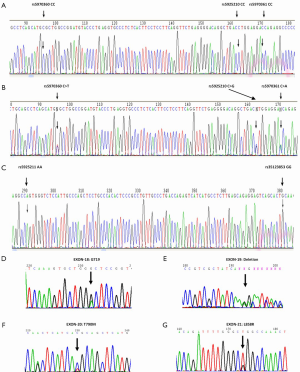
In all 191 tumors, two cases were detected with EGFR exon 18 point mutations (Figure 1D); 19 cases with exon 19 deletion mutations (Figure 1E); seven cases with exon 20 point mutations (Figure 1F); 20 cases with exon 21 point mutations (Figure 1G). A total of 46 cases had EGFR mutations of any subtype. Our analysis showed that the SNP loci genotypes were not correlated with EGFR mutation status or its subtypes (P>0.05; Table 1).
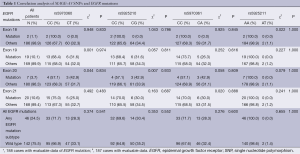
Full table
MAGE-A3 SNPs correlated with EGFR mRNA expression level
EGFR mRNA expression levels in the whole group averaged 3.85±0.48. EGFR mRNA expression levels of the SNP genotypes CC/CT on loci rs5970360 were 3.91±0.47 and 3.71±0.50, and the SNP genotypes CC/CG on loci rs5925210 were 3.91±0.47 and 3.73±0.49, and the SNP genotypes CC/CA on loci rs5970361 were 3.89±0.47 and 3.75±0.50, respectively. The genotypes of SNP loci rs5970361 was not correlated with the EGFR mRNA expression level. But the genotypes of SNP loci rs5970360 and rs5925210 were statistically correlated with the EGFR mRNA expression levels (Table 2).
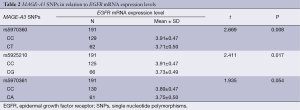
Full table
Correlation analysis of SNP polymorphism and clinical parameters
The genotypes of SNP loci rs5970360, rs5925210 and rs5970361 on MAGE-A3 gene were CC/CT, CC/CG and CC/CA, respectively. Our analysis showed that the SNP loci genotypes were not correlated with clinical parameters, such as gender, age, smoking status and pathological classification (P>0.05; Table 3).

Full table
Survival analysis
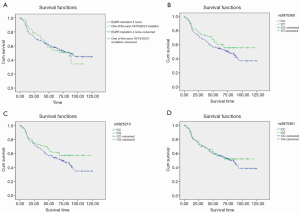
No significant differences were found between the EGFR mutation group and wild type group (56.4 vs. 59.8 months, P=0.838; Figure 2A), and among between the groups of SNP loci genotypes (56.4 vs. 64.8 months, P=0.185; Figure 2B, 56.4 vs. 65.1 months, P=0.127; Figure 2C, 58.8 vs. 52.2 months, P=0.648; Figure 2D).
Discussion
NSCLC was a malignancy that has the highest morbidity and mortality and the poor prognosis. Five-year survival of patients was only about 10-15%. The occurrence of tumor was a process that the control channel of normal cells adjustment and growth were influenced by all congenital and acquired factors (8,9). In recent years, more studies have shown that genetic factors has a close relation with lung cancer, especially the gene polymorphisms among people (10). MAGE-A3 gene is a member of the CT (cancer/testis) gene families and one of the most popular CT antigens which expressed in tumor tissue (11). MAGE-A3 antigen has become an active immunotherapy target in many malignant tumors including NSCLC and the clinical trial of treatment for early stage NSCLC with vaccine of MAGE-A3 protein was underway. But there was little report concerning whether MAGE-A3 gene polymorphism or mutations could affect individual specificity of MAGE-A3 gene expression and as the tumor prognosis factor to predict the survival of NSCLC patients.
Five SNP loci of MAGE-A3 gene in this research were located on the introns (rs5970360), exon 2 (rs5825210, rs5970361) and exon 3 (rs5925211, rs35123853), respectively. Our research shows that loci rs35123853A >G of Chinese NSCLC population were all GG genotype, its different from SNP database of NCBI which reported this loci has AA and AG two genotypes. The allele frequency of rs5925211A >T in Caucasian patient population also was reported in SNP database of NCBI. Allele frequency of rs5925211A >T was AA: 0.059, AT: 0.294 and TT: 0.647. But the genotypes we had found in Chinese NSCLC population were AA and AT. Allele frequency was 0.681 (CC, 130/191) and 0.319 (CT, 61/191) for the SNP loci rs5970360, and 0.660 (CC, 126/191) and 0.340 (CG, 65/191) for the SNP loci rs5925210, and 0.686 (CC, 131/191) and 0.314 (CA, 60/191) for the SNP loci rs5970361. NCBI haven’t reported the three loci. Genotype frequencies of SNP loci on MAGE-A3 in Chinese NSCLC population was significantly different from Caucasian patient population.
Ito et al. have reported the MAGE-A3 positive expression of the patients was the 50% (12). Most of the research showed that the expression level of CTAs has nothing to do with the gender of the patients (13,14), and MAGE-A gene was located on the X chromosome, only expressed in the human spermatogonium under normal circumstances (15), This reminded us that male patients with squamous carcinoma may have a high expression rate of MAGE-A3, and it has yet to be further improved in our subsequent research work.
Our study also found that no single factor correlation between three SNP loci genotype rs5970360, rs5825210 and rs5970361 and clinical parameters, such as gender, age, smoking status and pathological classification. Gene polymorphism of MAGE-A3 gene had no effect on survival of NSCLC patients, and these are consistent with reported by Ito (12). It is basically ascertained that it couldn’t be used for predicting the survival of NSCLC patients as the tumor prognosis factor.
EGFR mutation has been substantially established as a major molecular subtype of NSCLC, which can greatly benefit from EGFR tyrosine kinase inhibitors. In this study all fequencies of each MAGE-A3 SNPs were not found significantly different between EGFR mutant and wild type patients. But we found the genotypes of SNP loci rs5970360 and rs5925210 in MAGE-A3 had significant correlation with EGFR mRNA expression level. Thus MAGE-A3 SNPs might be predictive for EGFR expression levels, suggesting helpful to patient selection for EGFR protein target immunotherapy.
Limitations of this study include only SNPs of MAGE-A3. Methylation of MAGE-A3 may also influence its expression level.
In summary, here we report that allele frequencies of five SNP loci on MAGE-A3 gene in Chinese NSCLC population. Though SNPs of MAGE-A3 were not significantly associated with clinical factors, two major SNP loci rs5970360 and rs5925210 in MAGE-A3 had correlation with EGFR mRNA expression level. Thus genotyping of MAGE-A3 SNPs might be helpful to select patients for EGFR protein target immunotherapy. Prospective data and further investigations into molecular mechanisms underlying differential MAGE-A3 expression in tumors are warranted.
Acknowledgements
Funding: This study was supported by grants from the Foundation of Guangdong Science and Technology Department (Grant No. 2010B031600158, XN Yang) and Key Lab System Project of Guangdong Science and Technology Department (Grant No. 2012A061400006, YL WU).
Disclosure: The authors declare no conflict of interest.
References
- van der Bruggen P, Traversari C, Chomez P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991;254:1643-7. [PubMed]
- Kocher T, Schultz-Thater E, Gudat F, et al. Identification and intracellular location of MAGE-3 gene product. Cancer Res 1995;55:2236-9. [PubMed]
- Van den Eynde BJ, van der Bruggen P. T cell defined tumor antigens. Curr Opin Immunol 1997;9:684-93. [PubMed]
- Gure AO, Chua R, Williamson B, et al. Cancer-testis genes are coordinately expressed and are markers of poor outcome in non-small cell lung cancer. Clin Cancer Res 2005;11:8055-62. [PubMed]
- Weynants P, Lethé B, Brasseur F, et al. Expression of mage genes by non-small-cell lung carcinomas. Int J Cancer 1994;56:826-9. [PubMed]
- Brichard VG, Lejeune D. GSK's antigen-specific cancer immunotherapy programme: pilot results leading to Phase III clinical development. Vaccine 2007;25:B61-71. [PubMed]
- Yanagawa N, Tamura G, Oizumi H, et al. MAGE expressions mediated by demethylation of MAGE promoters induce progression of non-small cell lung cancer. Anticancer Res 2011;31:171-5. [PubMed]
- Ponder BA. Cancer genetics. Nature 2001;411:336-41. [PubMed]
- Sekido Y, Fong KM, Minna JD. Molecular genetics of lung cancer. Annu Rev Med 2003;54:73-87. [PubMed]
- Herbst RS, Onn A, Sandler A. Angiogenesis and lung cancer: prognostic and therapeutic implications. J Clin Oncol 2005;23:3243-56. [PubMed]
- Boël P, Wildmann C, Sensi ML, et al. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity 1995;2:167-75. [PubMed]
- Ito S, Kawano Y, Katakura H, et al. Expression of MAGE-D4, a novel MAGE family antigen, is correlated with tumor-cell proliferation of non-small cell lung cancer. Lung Cancer 2006;51:79-88. [PubMed]
- Peng JR, Chen HS, Mou DC, et al. Expression of cancer/testis (CT) antigens in Chinese hepatocellular carcinoma and its correlation with clinical parameters. Cancer Lett 2005;219:223-32. [PubMed]
- Nagorsen D, Scheibenbogen C, Marincola FM, et al. Natural T cell immunity against cancer. Clin Cancer Res 2003;9:4296-303. [PubMed]
- De Smet C, De Backer O, Faraoni I, et al. The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci U S A 1996;93:7149-53. [PubMed]


