Knockdown of HMGB1 improves apoptosis and suppresses proliferation and invasion of glioma cells
Introduction
A glioma is a primary brain tumor that originates from the nerve stromal cells including glial cells, ependymal cells, choroid plexus epithelial cells, and nerve parenchymal cells. Gliomas are featured by high morbidity, high recurrence rate, and high case-fatality rate (with an overall 5-year survival of only 13%), and low cure rate. Gliomas account for about 60% of all intracranial primary brain tumors, posing serious threat to the life and health of patients (1,2). Tumor invasion and metastasis are important biological characteristics of malignant tumors. Most patients who die of cancer die from metastatic disease rather than from the primary lesion (3,4). Gliomas are highly invasive. They often invade the surrounding normal brain tissue, and have no clear boundaries with normal brain tissues. Thus, surgery has only a limited role in treatment, and the case-fatality rate of gliomas is extremely high (1,5,6).
Cancers are caused by the accumulation of genetic mutations, including the activation of oncogenes and the loss of tumor suppressor genes. Activation of oncogenes can lead to cell malignant transformation and tumorigenesis (7-9). Most studies have shown that the activation of oncogenes play key roles in the occurence and development of gliomas (10-12). Activation of oncogenes usually involves the changes in multiple signaling pathways, cell cycle, and regulation of apoptosis (13-16). Recent research has shown that the high mobility group protein box 1 (HMGB1) gene is highly expressed in various tumours, showing oncogene-like functions (17-19). Forty years ago, HMGB1 was discovered in calf thymus and named according to its electrophoretic mobility in polyacrylamide gels (20). Human HMGB1 is a structural transcription factor encoded by a single gene. Located in chromosome 13q12, it contains four introns and five exons, encoding a 215-amino-acid protein, with a molecular weight of about 30 KD. It is highly conserved, with an amino acid sequence homology of over 98% between human and rodents (21,22). HMGB1 protein is divided into three domains: two positively charged DNA-binding motifs (boxes A, B) and a C-terminal acidic tail. Both the two motifs contain 80-90 amino acid residues and are strongly alkaline; and the carboxy terminus, also known as acidic terminal, is rich in aspartic acid and glutamic acid containing negative charge (23). HMGB1 is a multifunctional protein. HMGB1 functions as a DNA-binding protein in the nucleus; by binding DNA with specific structures, it can affect the structural state of the target sequence and thus participates in the key life activities including the division, differentiation, and maturation of cells, DNA repair, DNA recombination, regulation by steroid hormones, and regulation of gene transcription (24-27). Outside the cells, HMGB1 released by necrotic cells or activated immune cells is a typical injury-related molecule, which, by interacting with cytokines, chemokines, and growth factors, is involved in the various activities of cells (20). HMGB1 protein, as a ubiquitous nuclear protein, is widely distributed in mammalian cells, in particular in thymus, lymph tissue, testis, and neonatal liver (28). As shown in recent research, high expression of HMGB1 has also been detected in immature cells and a variety of solid tumors. HMGB1 is an anti-apoptotic protein; its over-expression can suppress apoptosis and thus cause the occurrence and development of tumors (29). In addition, HMGB1 is also related with the activation of the plasminogen system and matrix metalloproteinases and the migration of adherent cells (30,31). HMGB1 has been found to be highly expressed in many malignancies including colon cancer, lung cancer, breast cancer, nasopharyngeal carcinoma, and head and neck squamous cell carcinoma (32-36), and may also be closely associated with the occurrence, invasion, and metastasis of tumors. However, the specific mechanisms governing the biological activities of HMGB1 remain unclear, and few studies have explored the role ofHMGB1 in the development of glioma cells. Thus, in our current study, we inhibited the expression of HMGB1 gene in U251 and U-87MG cell lines using the gene knockout method; then, we detected the change of the biological characteristics of this cell line to analyze the effect of HMGB1 on the biological behaviors of glioma cells and explore the relationship between HMGB1 and the development, invasion, and metastasis of glioma, with an attempt to provide scientific evidences for the prognosis and targeted therapy of gliomas.
Materials and methods
Materials
Trizol reagent and RT-PCR kit were purchased from BioRad Company (Richmond, CA, USA), HMGB1 siRNA oligonucleotides targeting human HMGB1, control oligonucleotides [HMGB1 siRNA negative control (NC)] and transfection reagents were from RiboBio (Guangzhou, China). Trypsin, methyl thiazolyl tetrazolium (MTT), propidium iodide (PI), and dimethyl sulfoxide (DMSO) were from Sigma Corp. (St. Louis, Missouri, USA), Annexin V-FITC was purchased from Beyotime (Haimen, China). Human HMGB1 (NM_002128.4) upstream primer (GGAGAGTAATGTTACAGAGCGG) and downstream primer (AGGATCTCCTTTGCCCATGT) were synthesized by Shanghai Biological Engineering Technology Services Limited (Shanghai, China). Matrigel was purchased from BD Bioscience (San Jose, CA, USA), and transwell invasion chamber was from Corning Corp. (Midland, Michigan, USA). Protein extraction and protein quantification kits were purchased from Bio-Rad (Richmond, CA, USA). Rabbit anti-HMGB1 and rabbit anti-Bax polyclonal antibodies were from Abcam (Cambridge, MA, USA). Rabbit anti-matrix metalloproteinase-9 (MMP-9) polyclonal antibody was from LifeSpan Biosciences (Seattle, WA, USA). Rabbit anti-Bcl-2 polyclonal antibody and mouse anti-cyclin D1 polyclonal antibody were from Biorbyt (Cambridge, UK). Rabbit anti-tubulin polyclonal antibody was from Abbiotec (San Diego, CA, USA). Horseradish peroxidase conjugated goat anti-rabbit or rabbit anti-mouse IgG polyclonal antibody was from Invitrogen-Life Technologies (Carlsbad, CA, USA). ECL chemoluminescence kit was from Pierce Company (Rockford, Illinois, USA).
U251 and U-87MG cell culture and HMGB1 siRNA transfection
Human glioma cell lines U251 and U-87MG were purchased from ATCC (Rockville, MD, USA). The U251 and U-87MG cells were cultured at 37 °C in an environment with saturated humidity and 5% CO2 in the Eagle’s minimum essential medium (Eagle’s MEM) (Rickmansworth, England) containing 10% fetal bovine serum (Gibco Company, Grand Island, NY, USA), penicillin (100 U/mL), and streptomycin (100 µg/mL). Cells in the exponential phase were selected for further experiment. One day before transfection, the U251 and U-87MG cells at the logarithmic phase were digested with trypsin and then counted. Cells were inoculated in 6-well plates at an appropriate density. On the transfection day, after the cells were 90% confluent, they were treated overnight with Eagle’s MEM without serum. According to manufacturer’s instruction, the U251 and U-87MG cells in the exponential phase were transfected with HMGB1 siRNA (inhibiting HMGB1 mRNA) and NC siRNA (scrambled the nucleotide sequence of HMGB1 siRNA lacked homology to any other gene). Three groups were set: HMGB1 siRNA group, NC siRNA group and blank control group (transfected with only transfection reagents). The transfection and inhibition rates of HMGB1 siRNA were detected using Western blotting.
Cell viability: analysis by MTT test
Cell viability following HMGB1 knockdown was assessed by MTT test. In the Eagle’s minimum essential medium (Eagle’s MEM) containing 10% FBS, the U251 and U-87MG cells were prepared into single cell suspensions and then inoculated in the 96-well plates with 2,000 cells/well in 200 µL. The wells were then cultured in an incubator (37 °C and 5% CO2) until monolayers were obtained, which were adherent 24 hours later. Add 100 µL MTT solution (0.5 mg/mL) in each well, and then put the plate into the incubator to allow the cell culture process to continue. After 4 hours of culture, the medium was removed and 150 µL DMSO was added to each well. Then plates were placed on shaker for 10 min to fully dissolve the crystals. The OD value was measured at a wavelength of 570 nm. With this procedure, only viable cells with functioning mitochondria can oxidize MTT to a violet-red reaction product.
Cell proliferation: analysis by flow cytometry
Cell proliferation following HMGB1 knockdown was assessed by flow cytometry. After the cells were 70-80% confluent, they were refed with serum-free medium for 24 hours; after have been synchronized, they were cultured in complete medium for another 24 hours. Cells were collected (about 1×106-5×106/mL) and centrifuged at 800 r/min for 5 min to discard the culture medium. After the cells were washed with 3 mL PBS once, the mixture was centrifuged to remove PBS solution, and then added with ice-cold 70% ethanol and fixed at 4 °C for 1-2 hours. Discard the fixative by centrifugation. Resuspend the cells in 3 mL PBS for 5 min. After filtering through the 400-mesh filter cloth, the mixture was centrifuged at 800 r/min for 5 min to discard the PBS. Add 1 mL of PI staining solution and incubate 30 minutes in the dark at 4 °C. Detection using flow cytometer: for PI, cells were excited with an Argon 488 nm laser and emission of radiation at wavelengths of larger than 630 nm. Fluorescence intensity histogram profiles of the PI were analyzed using red fluorescence. For cell cycle analysis, data were expressed as fractions of cells in different cycle phases. Measurements repeated three times in each group.
Detection of cell apoptosis by flow cytometry
Apoptosis was assayed using the Annexin V-FITC Apoptosis Kit (Haimen, China) according to the manufacturer’s instructions. Briefly, after the cell culture medium was dried, the adherent cells were washed with PBS once, and then a proper amount of trypsin-EDTA solution was added to digest the cells. The cells were further inoculated at room temperature until they could be dispersed with gentle pipetting. Then, the trypsin-EDTA solution was removed. Add cell culture medium again, and then the mixture was transferred to a centrifuge tube. Centrifuge the cells at 1,000 rpm for 5 min. After the supernatant was discarded, add PBS and gently resuspended cells. Then, the cells were counted. Collect 50,000-100,000 resuspended cells, centrifuge them at 1,000 rpm for 5 min, and then discard the supernatant. A total of 195 µL annexin V-FITC liquid and 5 µL annexin V-FITC were added gently to the suspended cells, mixed gently, and incubated in room temperature (20-25 °C) at dark for 10 minutes (aluminum foil can be used to avoid light). Centrifuge for 5 minutes, and then the supernatant was discarded. A total of 190 µL annexin V-FITC liquid was added gently to the suspended cells. A total of 10 µL of PI staining solution was added, gently mixed, and incubated in ice bath at dark. Samples were analyzed by flow cytometry. Stain the cells simultaneously with FITC-Annexin V (green fluorescence) and PI (red fluorescence). Data were analyzed using Cell Quest software. Measurements repeated three times in each group.
Determination of invasion of glioma cells
The invasive ability of U251 and U-87MG glioma cells was assessed through the number of cells passed through an 8-µm pore size polycarbonate filter coated with Matrigel (Corning, MA, USA). The chamber was washed with serum-free medium three times. Add 20 µL appropriately diluted Matrigel, which was evenly covered on the surface of polycarbonate membrane. Inoculate at 37 °C for 30 min to create an artificial basement membrane. Thus, the chamber was divided into upper and lower chambers. In each group (HMGB1 siRNA group, NC siRNA group, and blank control group), the cells (2×105 cells) were suspended in 200 µL serum-free Eagle’s MEM medium, and then inoculated in the upper chamber of the transwell invasion system. Add 500 µL Eagle’s MEM medium containing 10% FBS, put the cells in the incubator, and then culture them for 24 hours. Take out the upper chamber, remove the cells on the surface of the upper chamber using sterile cotton swab and stain the cells that had invaded the lower surface of the basement membrane using crystal violet. Count the number of cells that had passed through the transwell polycarbonate membrane using Leica microscope (Leica Company, Wetzlar, Germany). Eight visual fields were randomly observed for each sample, and the experiment was repeated three times.
RNA isolation and real-time PCR
Total RNA was extracted from U251 and U-87MG glioma cells with Trizol reagent as per the manufacturer’s protocol and subjected to reverse transcription using a reverse transcription primer. Real-time PCR reactions were performed using the iTaq Fast SYBR Green Supermix (BioRad) exactly following the manufacturer’s instructions and Applied Biosystems 7500 real-time PCR system. Relative quantification was performed by using the comparative Ct method and all results were compared to the control samples after normalizing to an endogenous control [Glyceraldehyde-3-phosphate dehydrogenase (GAPDH)]. Values are expressed as mean percent ± SE.
Western blotting
The total proteins were extracted using protein extraction kit, and the protein concentration was analyzed using a BCA assay kit (Bio-Rad, Richmond, CA, USA). The total proteins of 40 µg per hole were subject to SDS-PAGE gel electrophoresis. After the protein had been transferred onto a PVDF membrane (Millipore Company, Bedford, MA, USA), they were blocked in TBS solution (10 mM Tris-HCl, pH 7.5, 150 mM NaCl) containing 5% skimmed milk powder at room temperature for 1 h, then added with cyclin D1, Bax, Bcl-2, and MMP-9 primary antibody and incubated overnight at 4 °C. Wash 3 times with fresh TBST for 5 minutes. Add HRP-labeled goat anti-rabbit IgG or rabbit anti-mouse polyclonal antibody and inoculate at 37 °C for 1 h. Wash 3 times with fresh TBST for 5 minutes. Autoradiography was conducted using ECL chemiluminescent substrate. The change of its relative expression was analyzed using PDQuest software (Bio-Rad, Richmond, CA, USA).
Statistical analysis
The experimental data were analyzed using SPSS 17.0 software. Inter-group differences were analyzed by t-test and multiple comparisons among several groups were conducted by analysis of variance. P<0.05 was considered significantly different.
Results
High expression of HMGB1 in glioma cell lines
Real-time PCR and western blotting showed that, compared with the immortalized human astrocytes (SVG-p12 cell line), the expressions of HMGB1 mRNA and protein significantly increased in glioma cell lines (U251, U-87MG, and LN-18) (P<0.05) (Figure 1A-C). U251 and U-87MG cells were selected for further study. We inhibited the HMGB1 expression in U251 and U-87MG cells by RNAi technique. As shown in Western blotting, the expression of HMGB1 in U251 and U-87MG cells was significantly inhibited in HMGB1 siRNA group but showed no significant difference in the NC siRNA group and blank control group (Figure 1D,E). Thus, the HMGB1-knockout cell model was successfully established.
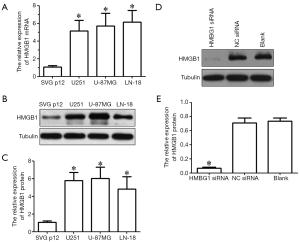
Effect of HMGB1 gene on the viability of glioma cells
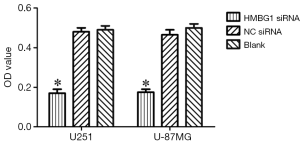
After the inoculation of equal volume of U251 and U-87MG cells in HMGB1 siRNA group, NC siRNA group, and blank control group, MTT assay showed that the viability of U251 and U-87MG cells was significantly lower in the HMGB1 siRNA group than in the NC siRNA group and blank control group (P<0.05) but was not significantly changed in the NC siRNA group and blank control group (Figure 2). Thus, the knockdown of HMGB1 gene may lower the viability of U251 and U-87MG cells, whereas the overexpression of HMGB1 gene may be associated with the increased vitality of U251 and U-87MG cells.
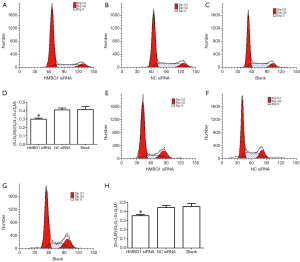
Effect of HMGB1 on the proliferation of glioma cells
After the inoculation of equal volume of U251 and U-87MG cells in HMGB1 siRNA group, NC siRNA group, and blank control group, flow cytometry after PI staining showed that the number of U251 and U-87MG cells in G0/G1 phases were significantly larger in the HMGB1 siRNA group than in the NC siRNA group and blank control group (P<0.05), whereas the numbers of cells in S phase and G2/M phases were significantly smaller (P<0.05); in contrast, the numbers of U251 and U-87MG cells in G1 phase, S phase, and G2/M phase showed no significant change in the NC siRNA group and the blank control group (Figure 3). Thus, the changes in the expression profile of HMGB1 gene may be related with the changes in the cell cycle of U251 and U-87MG glioma cells. More specifically, the suppressed expression of HMGB1 gene may be related with the cell-cycle G0/G1 phase arrest in U251 and U-87MG cells; or, the proliferation of U251 and U-87MG cells can be suppressed by inhibiting the expression of HMGB1 gene.
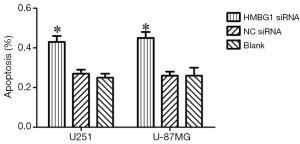
Effect of HMGB1 on Apoptosis of glioma cells
As shown by annexin V and PI staining, the HMGB1 siRNA group had significantly larger number of apoptotic U251 and U-87MG cells than the NC siRNA group and blank control group (P<0.05); however, no significant difference was found between the NC siRNA group and blank control group (P<0.05) (Figure 4). Therefore, the HMGB1 gene knockdown may inhibit the apoptosis of U251 and U-87MG cells.
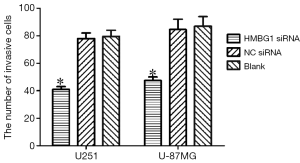
Effect of HMGB1 gene on the invasion ability of glioma cells
The invasive ability of U251 and U-87MG glioma cells was assessed through the number of cells passed through an 8-µm pore size polycarbonate filter coated with matrigel. As shown by the transwell invasion chamber assay, the number of cells passed through the polycarbonate filter was significantly smaller in the HMGB1 siRNA group than in the NC siRNA group and blank control group (P<0.05); however, no such different was found between the NC siRNA group and the blank control group (Figure 5). Therefore, the change of the HMGB1 protein expression level may be associated with the change of the invasion ability of U251 and U-87MG glioma cells. Knockout of HMGB1 gene may inhibit the invasiveness of U251 and U-87MG cells.
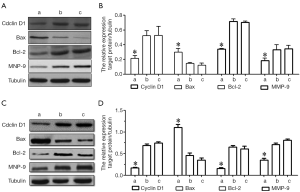
Effects of HMGB1 on cyclin D1, Bax, Bcl-2 and MMP-9 protein expressions
As shown by Western blotting, the HMGB1 siRNA group had significantly lower expression levels of cyclin D1, Bcl-2 and MMP-9 proteins and higher Bax protein expression level than the NC siRNA group and the blank control group (P<0.05); however, the protein expression profiles of cyclin D1, Bax, Bcl-2, and MMP-9 in U251 and U-87MG cells were not significantly changed in the NC siRNA group and the blank control group (Figure 6). Thus, the suppressed expression of HMGB1 may be associated with the down-regulated expressions of cyclin D1, Bcl-2, and MMP-9 protein and the up-regulated expression of Bax protein.
Discussion
HMGB1 is widely distributed in all mammalian tissues. The HMGB1 inside the nucleus and outside the cells exert different functions and biological effects. By binding DNA with specific structures, HMGB1 inside the nucleus can affect the structural state of the target sequence and thus participates in the key life activities including the division, differentiation, and maturation of cells, DNA repair, DNA recombination, regulation by steroid hormones, and regulation of gene transcription (24,25). In contrast, HMGB1 outside the cells can bind multiple receptors and thus exert different functions. It has been widely recognized that the main extracellular HMGB1 receptors include Toll-like receptors (TLRs) and receptor for advanced glycation end products (RAGE). The biological effects of HMGB1 are realized primarily through the interactions between RAGE and TLRs (37). Currently, the biological research on HMGB1 is mainly focused on the late inflammatory mediators, autoimmune diseases, axon growth, and tumor development and metastasis (25,33,38).
HMGB1 is a typical damage-associated molecular pattern (DAMP). Recent studies have demonstrated that HMGB1 is associated with the progression, invasion, and metastasis of tumors and has abundant expressions in a variety of tumor tissues and immature cells (39). HMGB1 is not only an important cytokine (40) but also closely related with tumors (41). HMGB1 has been found to be highly expressed in many malignancies including colon cancer, pancreatic cancer, breast cancer, nasopharyngeal carcinoma, and head and neck squamous cell carcinoma (38). Specific inhibition of the HMGB1 expression can suppress the malignant changes including the proliferation and invasion of tumor cells. However, few studies have reported the relationship between HMGB1 and the development of gliomas. In our current in vitro study, we tried to explore the role of HMGB1 gene in the U251 and U-87MG cells.
First, real-time PCR and Western blotting demonstrated the high expressions of HMGB1 mRNA and HMGB1 protein in glioma cell lines (U251, U-87MG, and LN-18), which was consistent with the previous reports on other tumors (32-36,42), suggesting that the up-regulated HMGB1 expression plays a key role in the development of gliomas. Thus, we further established a HMGB1-knockout cell model to investigate the role of HMGB1 in glioma cells. After the expression of HMGB1 in U251 and U-87MG cells was inhibited using the RNAi technique, the expression of HMGB1 in the U251 and U-87MG cells in the HMGB1 siRNA group was significantly suppressed, indicating that the HMGB1-knockout cell model was successfully established. Then, we explored the relationship between the changes in the HMGB1 expression and the biological characteristics of U251 and U-87MG cells. By using MTT method and PI staining, we detected the influences of HMGB1 gene on the growth, proliferation, and cell cycle of U251 and U-87MG cells. MTT assay indicated that the knockdown of HMGB1 gene may be associated with the decreased viability of U251 and U-87MG cells. PI staining showed that the decrease in the HMGB1 gene expression may be related with the G0/G1 arrest of U251 and U-87MG cells. These findings might be explained by the speculation that the expressions of the growth-promoting factors in the downstream signal transduction pathways of HMGB1 gene decrease and/or the growth-inhibiting factors increase. Therefore, we further detected the expression of cyclin D1, a positive regulator of the cell cycle (43) and found that the expression of cyclin D1 was significantly inhibited in the HMGB1 siRNA group. It is then speculated that the cell cycle of U251 and U-87MG cells in the HMGB1 siRNA group was blocked, which may be caused by the decreased expression of cyclin D1 protein; however, the exact mechanism remains unclear. We further used annexin V-FITC staining to investigate the effect of HMGB1 on apoptosis in U251 and U-87MG cells, and the results showed that the number of apoptotic U251 and U-87MG cells were significantly larger in the HMGB1 siRNA group than in the blank control group and NC siRNA group, suggesting that the decreased expression of HMGB1 might promote the apoptosis of U251 and U-87MG cells. It was found that the HMGB1 in Lewis cells could inhibit the expression of pro-apoptosis protein Bax and promote the expression of anti-apoptosis protein Bcl-2 (29). Was there any similar regulation pattern in the U251 and U-87MG cells? To answer this question, we detected the expressions of Bax and Bcl-2 and found that the expression level of anti-apoptosis protein Bcl-2 significantly decreased and the pro-apoptosis protein Bax protein level significantly increased in U251 and U-87MG cells in the HMGB1 siRNA group, which may explain the obvious apoptosis in U251 and U-87MG cells in the HMGB1 siRNA group. Some authors also found that the overexpression of HMGB1 could inhibit the activities of caspase-9 and caspase-3, suggesting that HMGB1 could inhibit the apoptosis (34). Thus, the inhibition of HMGB1 gene expression may inhibit the growth and proliferation of glioma cells and promote the apoptosis; overexpression of HMGB1 may promote the growth and proliferation of glioma cells and inhibit the apoptosis.
Recent studies have also found that the expression of HMGB1 is closely associated with the invasive growth of tumors, formation of metastatic lesions, and the poor prognosis (18,19,44-46). Thus, HMGB1 also plays a key role during tumor invasion and metastasis. In our current study, we used transwell invasion chamber assay to explore the effect of HMGB1 gene on the invasion ability of U251 and U-87MG cells and found that the number of U251 and U-87MG cells passed through the polycarbonate filter was significantly smaller in the HMGB1 siRNA group than in the NC siRNA group and blank control group, suggesting that the change in the HMGB1 protein expression level may be related with the invasion ability of U251 and U-87MG cells; in other words, knockout of HMGB1 gene may inhibit the invasiveness of U251 and U-87MG cells. HMGB1 can bind with its ligands RAGE and TLR and then induce the growth and migration of cells via its intracellular signaling pathways including NF-κB, MAPK, and ERK (47,48). It has been reported that the knockdown of HMGB1 by siRNA inhibited cell invasive potential, and down regulated the expression of NF-κB p65 and MMP-9 in gastric cancer cells (19). Exogenous HMGB1 induced the rapid phosphorylation of NF-κB and up regulated the expression of MMP-9 (49). Many other studies found that the changes in the expression of MMP-9 were significantly associated with the cell-matrix adhesion capacity (50-52). In addition, MMP-9 was found to be closely associated with tumor infiltration and metastasis (53-56). In our current study, we further explored the effect of HMGB1 on the expression of MMP-9 in U251 and U-87MG cells. We found that the expression of MMP-9 protein was significantly inhibited in the U251 and U-87MG cells in the HMGB1 siRNA group, indicating that the HMGB1 knockdown may down-regulate the expression of MMP-9 in U251 and U-87MG cells and thus exert its function in suppressing tumor infiltration and metastasis. However, its exact mechanism requires further study.
Conclusions
In conclusion, the up-regulated expression of HMGB1 may play key roles in the occurrence, development, invasion and metastasis of gliomas. Further understanding of the specific mechanisms via which HMGB1 regulates the development of gliomas will shed light on the new strategies and new targets for treating gliomas.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Rees JH. Diagnosis and treatment in neuro-oncology: an oncological perspective. Br J Radiol 2011;84:S82-9. [PubMed]
- Osman MA. Phase II trial of temozolomide and reirradiation using conformal 3D-radiotherapy in recurrent brain gliomas. Ann Transl Med 2014;2:44. [PubMed]
- Al-Mulla F, Bitar MS, Al-Maghrebi M, et al. Raf kinase inhibitor protein RKIP enhances signaling by glycogen synthase kinase-3β. Cancer Res 2011;71:1334-43. [PubMed]
- Wang J, Ma Y, Cooper MK. Cancer stem cells in glioma: challenges and opportunities. Transl Cancer Res 2013;2:429-41. [PubMed]
- Cai LB, Li J, Lai MY, et al. Bevacizumab rescue therapy extends the survival in patients with recurrent malignant glioma. Chin J Cancer Res 2013;25:206-11. [PubMed]
- Chen Y, Liu D. Chimeric antigen receptor (CAR)-directed adoptive immunotherapy: a new era in targeted cancer therapy. Stem Cell Investigation 2014;1:2.
- Ngalame NN, Tokar EJ, Person RJ, et al. Aberrant microRNA expression likely controls RAS oncogene activation during malignant transformation of human prostate epithelial and stem cells by arsenic. Toxicol Sci 2014;138:268-77. [PubMed]
- Shrestha Y, Schafer EJ, Boehm JS, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene 2012;31:3397-408. [PubMed]
- Baskaran D, Spirin PV, Prasolov VS. Activated leukemic oncogenes responsible for neoplastic transformation of hematopoietic cells. Mol Biol (Mosk) 2010;44:418-30. [PubMed]
- Boccaccio C, Comoglio PM. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res 2013;73:3193-9. [PubMed]
- Li J, Gupta S, Li C. Research perspectives: gold nanoparticles in cancer theranostics. Quant Imaging Med Surg 2013;3:284-91. [PubMed]
- Feng H, Hu B, Vuori K, et al. EGFRvIII stimulates glioma growth and invasion through PKA-dependent serine phosphorylation of Dock180. Oncogene 2014;33:2504-12. [PubMed]
- Ibrahim NY, Abdel Aal HH, Abdel Kader MS, et al. Reducing late effects of radiotherapy in average risk medulloblastoma. Chin Clin Oncol 2014;3:3.
- Paul AG, Chandran B, Sharma-Walia N. Cyclooxygenase-2-prostaglandin E2-eicosanoid receptor inflammatory axis: a key player in Kaposi's sarcoma-associated herpes virus associated malignancies. Transl Res 2013;162:77-92. [PubMed]
- McCubrey JA, Demidenko ZN. Recent discoveries in the cycling, growing and aging of the p53 field. Aging (Albany NY) 2012;4:887-93. [PubMed]
- Gandarillas A. The mysterious human epidermal cell cycle, or an oncogene-induced differentiation checkpoint. Cell Cycle 2012;11:4507-16. [PubMed]
- Süren D, Yıldırım M, Demirpençe Ö, et al. The role of high mobility group box 1 (HMGB1) in colorectal cancer. Med Sci Monit 2014;20:530-7. [PubMed]
- Xiao J, Ding Y, Huang J, et al. The association of HMGB1 gene with the prognosis of HCC. PLoS One 2014;9:e89097. [PubMed]
- Zhang J, Kou YB, Zhu JS, et al. Knockdown of HMGB1 inhibits growth and invasion of gastric cancer cells through the NF-κB pathway in vitro and in vivo. Int J Oncol 2014;44:1268-76. [PubMed]
- Kang R, Zhang Q, Zeh HJ 3rd, et al. HMGB1 in cancer: good, bad, or both? Clin Cancer Res 2013;19:4046-57. [PubMed]
- Riuzzi F, Sorci G, Donato R. RAGE expression in rhabdomyosarcoma cells results in myogenic differentiation and reduced proliferation, migration, invasiveness, and tumor growth. Am J Pathol 2007;171:947-61. [PubMed]
- Ditsworth D, Zong WX, Thompson CB. Activation of poly (ADP)-ribose polymerase (PARP-1) induces release of the pro-inflammatory mediator HMGB1 from the nucleus. J Biol Chem 2007;282:17845-54. [PubMed]
- Zhang CL, Shu MG, Qi HW, et al. Inhibition of tumor angiogenesis by HMGB1 A box peptide. Med Hypotheses 2008;70:343-5. [PubMed]
- Dong Xda E, Ito N, Lotze MT, et al. High mobility group box I (HMGB1) release from tumor cells after treatment: implications for development of targeted chemoimmunotherapy. J Immunother 2007;30:596-606. [PubMed]
- Tang D, Kang R, Zeh HJ 3rd, et al. High-mobility group box 1 and cancer. Biochim Biophys Acta 2010;1799:131-40.
- Bustin M. Revised nomenclature for high mobility group (HMG) chromosomal proteins. Trends Biochem Sci 2001;26:152-3. [PubMed]
- Bonaldi T, Längst G, Strohner R, et al. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J 2002;21:6865-73. [PubMed]
- Mosevitsky MI, Novitskaya VA, Iogannsen MG, et al. Tissue specificity of nucleo-cytoplasmic distribution of HMG1 and HMG2 proteins and their probable functions. Eur J Biochem 1989;185:303-10. [PubMed]
- Xu X, Zhu H, Wang T, et al. Exogenous high-mobility group box 1 inhibits apoptosis and promotes the proliferation of lewis cells via RAGE/TLR4-dependent signal pathways. Scand J Immunol 2014;79:386-94. [PubMed]
- Gnanasekar M, Kalyanasundaram R, Zheng G, et al. HMGB1: A Promising Therapeutic Target for Prostate Cancer. Prostate Cancer 2013;2013:157103.
- Carvajal IM, Baron RM, Perrella MA. High-mobility group-I/Y proteins: Potential role in the pathophysiology of critical illnesses. Crit Care Med 2002;30:S36-42.
- Akaike H, Kono K, Sugai H, et al. Expression of high mobility group box chromosomal protein-1 (HMGB-1) in gastric cancer. Anticancer Res 2007;27:449-57. [PubMed]
- Thomas JO, Travers AA. HMG1 and 2, and related 'architectural' DNA-binding proteins. Trends Biochem Sci 2001;26:167-74. [PubMed]
- Völp K, Brezniceanu ML, Bösser S, et al. Increased expression of high mobility group box 1 (HMGB1) is associated with an elevated level of the antiapoptotic c-IAP2 protein in human colon carcinomas. Gut 2006;55:234-42. [PubMed]
- Ranganathan A, Gunnarsson O, Casarett D. Palliative care and advance care planning for patients with advanced malignancies. Ann Palliat Med 2014;3:144-9.
- Lam S, Lin Y, Warnke PC. Permeability imaging in pediatric brain tumors. Transl Pediatr 2014;3:218-28.
- Apetoh L, Ghiringhelli F, Tesniere A, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med 2007;13:1050-9. [PubMed]
- Moriwaka Y, Luo Y, Ohmori H, et al. HMGB1 attenuates anti-metastatic defense of the lymph nodes in colorectal cancer. Pathobiology 2010;77:17-23. [PubMed]
- Fukami A, Adachi H, Yamagishi S, et al. Factors associated with serum high mobility group box 1 (HMGB1) levels in a general population. Metabolism 2009;58:1688-93. [PubMed]
- Yang H, Tracey KJ. Targeting HMGB1 in inflammation. Biochim Biophys Acta 2010;1799:149-56.
- Taguchi A, Blood DC, del Toro G, et al. Blockade of RAGE-amphoterin signalling suppresses tumour growth and metastases. Nature 2000;405:354-60. [PubMed]
- Chalmers SA, Eidelman AS, Ewer JC, et al. A role for HMGB1, HSP60 and Myd88 in growth of murine mammary carcinoma in vitro. Cell Immunol 2013;282:136-45. [PubMed]
- Atadja P, Wong H, Veillete C, et al. Overexpression of cyclin D1 blocks proliferation of normal diploid fibroblasts. Exp Cell Res 1995;217:205-16. [PubMed]
- Chen RC, Yi PP, Zhou RR, et al. The role of HMGB1-RAGE axis in migration and invasion of hepatocellular carcinoma cell lines. Mol Cell Biochem 2014;390:271-80. [PubMed]
- Yin Y, Li W, Deng M, et al. Extracellular high mobility group box chromosomal protein 1 promotes drug resistance by increasing the expression of P-glycoprotein expression in gastric adenocarcinoma cells. Mol Med Rep 2014;9:1439-43. [PubMed]
- Bald T, Quast T, Landsberg J, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature 2014;507:109-13. [PubMed]
- Fiuza C, Bustin M, Talwar S, et al. Inflammation-promoting activity of HMGB1 on human microvascular endothelial cells. Blood 2003;101:2652-60. [PubMed]
- Treutiger CJ, Mullins GE, Johansson AS, et al. High mobility group 1 B-box mediates activation of human endothelium. J Intern Med 2003;254:375-85. [PubMed]
- Gong W, Wang ZY, Chen GX, et al. Invasion potential of H22 hepatocarcinoma cells is increased by HMGB1-induced tumor NF-κB signaling via initiation of HSP70. Oncol Rep 2013;30:1249-56. [PubMed]
- Das SK, Bhutia SK, Kegelman TP, et al. MDA-9/syntenin: a positive gatekeeper of melanoma metastasis. Front Biosci (Landmark Ed) 2012;17:1-15. [PubMed]
- Chetty C, Vanamala SK, Gondi CS, et al. MMP-9 induces CD44 cleavage and CD44 mediated cell migration in glioblastoma xenograft cells. Cell Signal 2012;24:549-59. [PubMed]
- Sen T, Chatterjee A. Epigallocatechin-3-gallate (EGCG) downregulates EGF-induced MMP-9 in breast cancer cells: involvement of integrin receptor α5β1 in the process. Eur J Nutr 2011;50:465-78. [PubMed]
- Chen YY, Chiang SY, Lin JG, et al. Emodin, aloe-emodin and rhein inhibit migration and invasion in human tongue cancer SCC-4 cells through the inhibition of gene expression of matrix metalloproteinase-9. Int J Oncol 2010;36:1113-20. [PubMed]
- Kim A, Kim MJ, Yang Y, et al. Suppression of NF-kappaB activity by NDRG2 expression attenuates the invasive potential of highly malignant tumor cells. Carcinogenesis 2009;30:927-36. [PubMed]
- Kanno T, Kamba T, Yamasaki T, et al. JunB promotes cell invasion and angiogenesis in VHL-defective renal cell carcinoma. Oncogene 2012;31:3098-110. [PubMed]
- Argast GM, Krueger JS, Thomson S, et al. Inducible expression of TGFβ, snail and Zeb1 recapitulates EMT in vitro and in vivo in a NSCLC model. Clin Exp Metastasis 2011;28:593-614. [PubMed]


