Distribution and drug resistance of pathogenic bacteria isolated from cancer hospital in 2013
Introduction
To understand the characteristics of the pathogenic bacteria’s distribution and drug resistance, it was helpful to control and prevent the occurrence of drug-resistant bacteria. A lot of reports in general hospitals were searched with this drug resistance criterion. Lower immunity, longer admission time, and different cancers were characterized in addition to radiation, chemotherapy, and invasive diagnostic factors, such as operations for cancer patients which can lead to increased incidence of infections. In addition, to prevent infection, preventive and therapeutic using of high doses of antimicrobial agents easily lead to ecological imbalance endogenous flora, and multiple sites of infection in the tumor patients (1,2). According to the literature, mortality rate of tumor patients increased due to infectious diseases (3,4). Many data showed that the treatment efficiency significantly decrease in some tumor patients owing to infection (5). Therefore, it is very important to treat infection timely in tumor patients. So it was necessary to illustrate pathogenic bacteria distribution and drug resistance in order to provide the basis for antimicrobial agent therapy for tumor patients.
Materials and methods
Strains
A total of 1,378 strains of pathogenic bacteria were isolated from all types of specimens in all the departments of Peking University Cancer Hospital in 2013 (excluding the same repetitive strains from the same patient parts). The types of specimen included sputum, tracheal fluid, urine, blood, secretions, puncture drainage, fester, and so on.
Instruments and reagents
Pathogenic bacteria identification and drug susceptibility tests were performed with a VITEK 2 compact automatic identification system (bioMérieux Inc., Marcy I’Etoile, France). Fungal susceptibility was determined by ATB FUNGUS3 (bioMérieux Inc., Marcy I’Etoile, France). Drug susceptibility results refer to the standards (M100-S24) of Clinical and Laboratory Standards Institute (CLSI) in 2014.
Quality control strains
Staphylococcus aureus (ATCC 29213), Streptococcus pneumoniae (ATCC 49619), Escherichia coli (ATCC 25922), Enterobacter cloacae (ATCC 70032), and Candida albicans (ATCC 90028) were purchased from the quality control center of the ministry of health of China.
Statistical analysis
Data were analyzed using WHONET 5.6 software (World Health Organization Collaborating Centre for Surveillance of Antimicrobial Resistance).
Results
Pathogenic bacteria specimen distribution ratios in tumor patients
A total of 1,378 strains were detected from 4,530 specimens; among which 787 strains were isolated from the respiratory tract, accounting for 57.1%; 160 strains from blood, accounting for 11.6%; 89 strains from secretion, accounting for 6.5%; 71 strains from urine, accounting for 5.2%; 33 strains from puncture fluid, accounting for 2.4%; 32 strains from catheter, accounting for 2.3%; and 31 strains from fester, accounting for 2.2%.
Tumor pathogenic bacteria distribution
Of the 1,378 strains, 980 Gram-negative bacilli were detected, accounting for 71.1%, in which Klebsiella pneumonia, Escherichia coli and Pseudomonas aeruginosa were dominant. We found 328 Gram-positive coccus, accounting for 23.8%, in which the amount of Staphylococcus aureus was the highest. We identified 46 fungi, accounting for 4.1%, and 14 strains were anaerobic bacteria, accounting for 1.0% (Table 1).
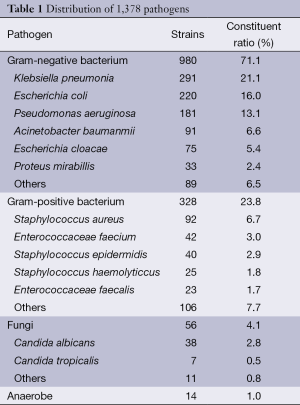
Full table
Department distribution
Of the 1,378 strains, the surgical departments detected 685 strains, accounting for 49.7%; the internal medicine departments detected 336 strains, accounting for 24.4%; the intensive care unit (ICU) detected 110 strains, accounting for 8.0%; the radiation department detected 95 strains, accounting for 6.9%; the interventional department detected 42 strains, accounting for 3.0%; and the Outpatient Service detected 110 strains, accounting for 8.0% (Table 2).
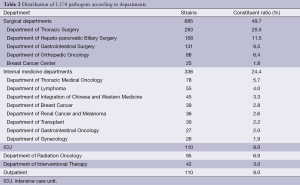
Full table
Cancer type distribution
Of the 1,378 strains, there were 288 cases of lung cancer, accounting for 20.9%; 238 cases of intestinal tumor, accounting for 17.3%; 195 cases of esophageal cancer, accounting for 14.2%; 157 cases of gastric cancer, accounting for 11.4%; 96 cases of lymphoma, accounting for 7.0%; and 54 cases of breast cancer, accounting for 3.9%, as shown in the Table 3.
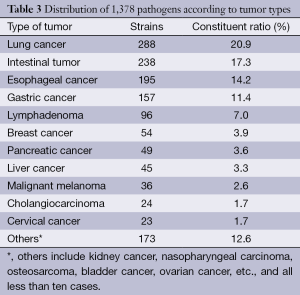
Full table
Drug resistance results
Common Gram-negative bacteria resistance rates
Forty-one of Klebsiella pneumoniae produced extend spectrum beta lactamase (ESBLs), accounting for 14.1%; and 150 of Escherichia coli produced ESBLs, accounting for 68.2%. Klebsiella pneumoniae and Escherichia coli were not resistant to imipenem and ertapenem. The resistance rates to imipenem in Pseudomonas aeruginosa and Acinetobacter baumannii were 14.9% and 13.2%, respectively (Table 4).
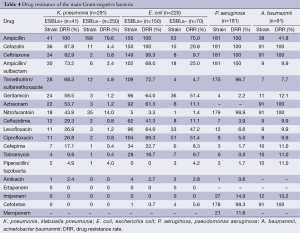
Full table
Common Gram-positive bacteria resistance rates
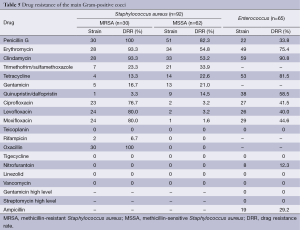
Staphylococcus aureus and Enterococcus were resistant to linezolid, but vancomycin resistance was not found (Table 5).
Full table
Fungal resistance rates
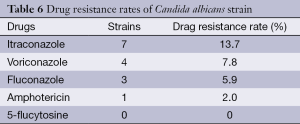
Itraconazol had the highest resistance percentage of 13.7% out of the 51 fungi (Table 6).
Full table
Discussion
Peking University Cancer Hospital is a specialized hospital. The patients admitted into this hospital have lower immune systems. High doses of multiple concurrent chemo-radiation can increase the chances of pathogenic bacteria invasion in most of the cancer patients hospitalized. As a result, nosocomial infections are sensitive to these types of patients. Moreover, radiation, chemotherapy, and long-term use of antibiotics lead to dysbacteriosis, which increases conditional pathogenic bacteria infections. Therefore, it is important to understand pathogenic bacteria and their drug resistance in patients with tumors for the initial clinical treatment experience and prognosis improvement.
Through the retrospective analysis of pathogenic bacteria performed in 2013, a total of 1,378 strains were detected. Respiratory specimens were the most prevalent (57.1%), followed by blood specimens (11.6%). Gram-negative bacteria and Gram-positive bacteria accounted for 71.1% and 23.8%, respectively, which was similar to the results of 2012 CHINET surveillance of bacterial resistance in China (6). The Gram-negative bacilli, Escherichia coli, Klebsiella pneumoniae, and Pseudomonas aeruginosa, were the top three bacteria detected, accounting for 21.1%, 16.0%, and 16.0%, respectively. The Gram-positive cocci, Staphylococcus aureus and Enterococcus faecium, were dominant, accounting for 6.7% and 3.0%, respectively. Li et al. reported that in Shandong Province Tumor Hospital, the top three detected gram-negative bacillus were Klebsiella pneumonia, Pseudomonas aeruginosa, and Escherichia coli, and the top two Gram-positive bacteria were Staphylococcus aureus and streptococcus pneumonia (7). We proposed that the different pathogenic spectrum may be due to different cities, different hospitals, different geographical locations, and different operations and diseases. This research showed that fungi accounted for 4.1% of all pathogenic bacteria, which is slightly lower than the domestic reports (7). Fourteen strains of anaerobic bacteria accounted for 1.0%, among which Bacteroides fragilis was dominant.
In this study, the highest proportion of pathogens detected was in the surgical departments, due to more surgical inspection specimens and infection possibilities caused by operations, invasive diagnosis and treatments. Chest surgery had the highest detection because the patients with lung cancer were prone to lower respiratory tract infections and respiratory specimen collections. The internal medicine departments had a lower detection rate. Our study also found that the detection of pathogenic bacteria in lung cancer had the highest rate (20.9%), which was caused by impaired lung function in patients, especially in patients with advanced disease who stay in bed for a long time. In addition, chemotherapy and radiation decreased respiratory cilia functions, which decrease the function of clearing secretions and defense functions. Detection of pathogenic bacteria in intestinal tumors was secondary, accounting for 17.3%. It most likely caused intestinal infection or obstruction through surgery or tumor. Other tumors, such as breast cancer, liver cancer, pancreatic cancer and melanoma, had a lower detection rate of less than 4%.
According to the analysis of the drug resistance of pathogenic bacteria isolated from the hospital, Klebsiella pneumoniae and Escherichia coli were the top two detected, which produced ESBLs with a rate of 14.1% and 68.2%, respectively. The separation rate of ESBLs-positive Klebsiella pneumoniae was lower than the domestic reports, while the separation rate of ESBLs-positive Escherichia coli was higher than the domestic reports (6,8,9). Because of ESBLs encoding gene on a plasmid, its resistance can be through transformation, transduction, or translocation in different bacteria. Therefore, ESBLs-producing bacteria are often easy to carry on multiple drug resistance (10). Our data indicated that ESBLs-positive Enterobacteriaceae had a higher drug resistance rate than ESBLs-negative bacteria. The resistance rates to gentamicin, ampicillin/sulbactum, sulfanilamide, aztreonam and cephalosporin, except for cefepime and ceftazidime, were greater than 50.0% in ESBLs-positive Klebsiella pneumonia. The resistance rates to fluoroquinolone gentamicin, sulfanilamide, aztreonam and cephalosporin, except for cefepime and ceftazidime, were also more than 50%. Thus, caution should be taken to these drugs for treatment. Carbapenem had the highest sensitivity in Enterobacteriaceae. Our hospital did not demonstrate imipenem and ertapenem resistance in Escherichia coli and Klebsiella pneumonia. Piperacillin/sulbactum, amikacin, and cefotetan had higher sensitivity, so that they could be used for experience therapy.
Pseudomonas aeruginosa and Acinetobacter baumannii were the highest detected non-fermentative bacteria. Pseudomonas aeruginosa virulence was stronger and more prone to induced resistance; therefore, great attention should be paid. Pseudomonas aeruginosa had more than 90% sensitivity to piperacillin/sulbactum, amikacin, tobramycin, cefepime, cephalosporin, gentamicin, and fluoroquinolone, The resistance rates to imipenem and meropenem were 11.6% and 14.9%, respectively, which were lower than domestic reports (11). Pakyz et al. reported that Pseudomonas aeruginosa nosocomial isolation had a significant linear relationship with carbapenem usage. Restriction of carbapenem usage would reduce drug resistance occurrence (12). In addition, for Pseudomonas aeruginosa infection treatment, early effective antimicrobial therapy is crucial. Combination antimicrobial agents could play into the synergy of drugs and reduce the occurrence of drug resistance (13). Nosocomial infections with Acinetobacter baumannii and Methicillin-resistant Staphylococcus aureus (MRSA) were due to iatrogenic factors (medical personnel, medical apparatus and instruments, etc.) (14). In recent years, imipenem-resistant Acinetobacter has grown (15). Acinetobacter baumannii resistance rates to imipenem were 13.2% in our hospital. The resistance to cefepime, piperacillin/sulbactum, fluoroquinolone, gentamicin, cefazolin, tobramycin and sulfanilamide were less than 20.0%. Therefore, the treatment of non-fermentative bacteria should be performed according to the result of drug susceptibility and the abuse of antimicrobial agents should be avoided.
We did not find Gram-positive resistance to vancomycin, teicoplanin, or linezolid, which can be used as the drug of choice for serious Staphylococcus infections. Staphylococcus had more than a 50% resistance rate to the traditional antibacterial drugs such as penicillin, erythromycin, and clindamycin. Clinics should be careful to use these drugs. It should be pointed out that vancomycin and linezolid-resistant Enterococcus faecalis and Enterococcus faecium were not found.
Acknowledgements
Bacteria with antimicrobial resistance had strong regional differences; there are even differences between the hospitals. This study retrospectively analyzed all the whole 2013 bacteria resistance data to provide a scientific basis for choosing the right antibiotics in order to reduce the morbidity and mortality of infection.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Song XY, Zhang L, Ye B, et al. Pathogenic distribution and antimicrobial resistance of gram-negative bacilli causing nosocomial infections in patients with tumor. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2012;22:838-9.
- Shen JW, Tian LH. Antimicrobial resistance of acinetobacter baumanii causing nosocomial infection in department of tumor. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2011;21:2828-9.
- Zhang JL, Ni MX, Ji QH, et al. Nosocomial infection in inpatients with cancer in tumor hospital:investigation and analysis. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2011;21:1334-6.
- Zhu B, Wang Y, Chen YH, et al. Nosocomial infection in patients with malignant tumor after radiotherapy and chemotherapy. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2010;20:2421-2.
- Li ZY, Liang Y, Zhang Q, et al. Investigation of nosocomial infection in 186 older tumor inpatients. Xian Dai Zhong Xi Yi Jie He Za Zhi 2010;159:1940-1.
- Wang F, Zhu DM, Hu FP, et al. 2012 CHINET surveillance of bacterial resistance in China. Zhongguo Gan Ran Yu Hua Liao Za Zhi 2013;13:321-30.
- Li DM, Song XR, Wang SP, et al. Distribution and drug resistance of pathogenic bacteria isolated from tumor hospital in 2012. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2014;24:2386-8.
- Xie JZ, Xue FY, Huang B, et al. Distribution and antibacterial resistance study on nosocomial infection pathogens in tumor patients. Zhongguo Yu Fang Yi Xue Za Zhi 2010;11:139-43.
- Li PZ, Chen YH, Lu YP, et al. Distribution of pathogens in tumor patients with nosocomial infection and their antibiotic resistance. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2008;18:1764-6.
- Zahar JR, Lesprit P. Management of multidrug resistant bacterial endemic. Med Mal Infect 2014;44:405-11. [PubMed]
- Cai H, Lin H. Analysis of distribution and antibiotic resistance of Pseudomonas aeruginosa isolated between 2008 and 2012. Zhongguo Shi Yan Zhen Duan Xue 2013;17:1666-8.
- Pakyz AL, Oinonen M, Polk RE. Relationship of carbapenem restriction in 22 university teaching hospital to carbapenem use and carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother 2009;53:1983-6. [PubMed]
- Wang J, Liang J, Xiao YH. Mohnarin of 2008: bacterial composing and resistance in bloodstream infections. Zhonghua Yi Yuan Gan Ran Xue Za Zhi 2010;20:2399-404.
- Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med 2008;358:1271-81. [PubMed]
- García-Garmendia JL, Ortiz-Leyba C, Garnacho-Montero J, et al. Risk factors for Acinetobacter baumannii nosocomial bacteremia in critically ill patients: a cohort study. Clin Infect Dis 2001;33:939-46. [PubMed]


