Genetic variants in RAN, DICER and HIWI of microRNA biogenesis genes and risk of cervical carcinoma in a Chinese population
Introduction
Cervical cancer is one of the most common malignancies among women worldwide, with an estimated 530,000 new cases and 275,000 deaths in 2008 (1), and 83% of the cases had occurred in developing countries (2). As a developing country, cervical cancer incidence and mortality rates have been substantial declines in the past 30 years, but 135,000 new cases are still diagnosed in China every year (3). Epidemiologic and laboratory-based studies indicated that the prevalence of genital human papillomavirus (HPV) infection in developing countries is very high in young females, but most of the infected can regress without intervention in two years, only a little high-risk HPV persistent infection might ultimately lead to cervical cancer (4). Given the mounting evidence that HPV infection is necessary but not sufficient cause for cervical cancer development, epidemiologic studies have suggested that host genetic variations may play an important contribution to cervical cancer susceptibility (5,6).
Recently discovered microRNAs (miRNAs) are a class of small noncoding RNAs, which are about 22 nucleotides in length, and modulate gene expression by targeting mRNA for deregulation or translational repression (7,8). These molecules control the expression of the majority of protein-encoding genes in humans. Therefore, miRNA appears to play critical roles either in physiological and pathological mechanisms, including cancer (9,10). A global deregulation of mature miRNA expression has been shown in several human cancers including cervical cancer, and specific miRNAs could be used to define molecular subtypes of different cancers (11). Along with these studies, it has been suggested that impaired miRNA processing could promote cellular transformation and tumorigenesis (12).
miRNA genes are initially transcribed by RNA polymerase II to form large primary miRNAs (pri-miRNAs). After transcription, pri-miRNAs are processed in the nucleus by the microprocessor machinery containing the RNase III Drosha and RNA binding protein DGCR8.This processing releases 60-70 nucleotide precursor miRNAs (pre-miRNAs), which are exported to the cytoplasm via Ran-GTPase and Exportin-5 (XPO5). Then pre-miRNAs are processed to produce the mature miRNAs through a protein complex that includes DICER, TRBP, AGO1, AGO2, HIWI, GEMIN3, GEMIN4, etc (7). In addition to deregulation of miRNAs, the aberrations of miRNA processing genes have been associated with several types of cancer. For example, altered expression of DICER modified the development of lung cancer, breast cancer, colorectal cancer and prostate cancers (13-16). Li et al. found that HIWI was universally upregulated in many types of cancer including cervical cancer and played an important role in oncogenesis (17). Given the critical function of miRNA biogenesis and the involvement of cancer development and progression, it is not surprising that genetic variations in miRNA biogenesis pathway could affect risk of cancers.
Single nucleotide polymorphisms (SNPs) are the most common human genetic variants and may contribute to an individual’s susceptibility to cancer. At present, multiple studies have revealed that the presence of SNPs in miRNA biogenesis pathway genes as well as miRNA genes and their target binding sites could affect the risk of cancer development, treatment response, and patient survival (18). Genetic variants in miRNA biogenesis pathway genes have been reported to be associated with the risk of several cancers such as bladder cancer, renal cell carcinoma, lung cancer, esophageal cancer, and breast cancer (19-23). However, few studies had shown the association between the polymorphisms in the miRNA biogenesis pathway genes and the risk of cervical cancer. Therefore, we hypothesized that SNPs in the miRNA processing genes may influence their expression or activity, thereby modulating susceptibility to cervical cancer. To test this hypothesis, we conducted a case-control study to evaluate the associations between SNPs in the miRNA processing genes (rs1057035 in DICER1, rs10773771 in HIWI and rs3803012 in RAN) and cervical cancer risk.
Materials and methods
Study population
The case-control study was approved by the Institutional Review Board of Nanjing Medical University. A total of 1,486 cervical cancer cases and 1,549 cancer-free controls were included in this study. All subjects were genetically unrelated Han Chinese women, and informed consents of approximately 95% of them were obtained. In brief, cervical cancer cases were recruited from the First Affiliated Hospital of Nanjing Medical University and the Tumor Hospital of Nantong City, Jiangsu, China, from March 2006 to January 2010. All cervical cancer cases in the study were newly diagnosed and histologically confirmed without restrictions of age or histological type in the two hospitals. Cancer-free women controls were randomly selected from a cohort of more than 30,000 participants in a community-based screening program for non-infectious diseases conducted in Jiangsu Province during the same time period as the cases were recruited. These control subjects had no self-reported cancer history and were frequency matched to the cases on age (±5 years) and residential areas (urban and rural). Each individual was interviewed by trained interviewers so as to collect information on demographic data, menstrual and reproduction history, and environment exposure history. After an informed consent, 5 mL of venous blood sample was collected from each subject for later genotyping assays.
SNP selection and genotyping
Based on the databases of the International HapMap Project (http://www.hapmap.org), dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP/) and bioinformatics prediction (http://snpinfo.niehs.nih.gov/snpfunc.htm), we searched the potentially functional polymorphisms (MAF >0.05) in RAN, DICER and HIWI genes for Asians. As a result, we selected three common SNPs located at 3'-untranslated regions (UTR): rs3803012 (A > G) in RAN, rs1057035 (C > T) in DICER and rs10773771 (C > T) in HIWI.
Genomic DNA was extracted from leukocytes of venous blood by proteinase K digestion and was followed by phenol-chloroform extraction and ethanol precipitation. The genotyping of the SNPs was performed by using TaqMan allelic discrimination Assay (primer: sense, 5'-TTTG GAACGCAGTTGATTCCT-3', antisense, 5'-GTAAC AAACAAGATTTCACCAC TGAATAT-3', probe: allele A, FAM-AGTTTCATATA TAAGACTGC, allele G, HEX-AGTTTCATATGTA AGACTG for rs3803012; primer: sense, 5'-TCTGCAGTTGCTTTTTCAAGACA-3', antisense, 5'-GAGACCG AATGTAATATGGAAAACCT-3', probe: allele C, FAM-TACACACGCGCTCAG, allele T, HEX-TACACACGTGCTCAGG for rs1057035; primer: sense, 5'-GTATCTTATCCA CATTTCTTC-3', antisense, 5'-TTCTAGCATTGCTATTCAC-3', probe: allele C, FAM-ACGTATAA AATAAGGAAGC, allele T, HEX-TACAT ATAAAATAAGGAAGC for rs10773771) on ABI PRISM 7900 HT platform (Applied Biosystems, Inc.). All the genotyping assays were performed without knowing the subjects’ case or control status, and approximately equal numbers of case and control samples were assayed in each 384-well plate with two blank controls. We also randomly selected 96 samples to genotype in duplicate for all the SNPs, and the results were 100% concordant.
Statistical analysis
We used χ2 test and student t-test to analyze the differences in demographic characteristics, selected variables and frequencies of the genotypes between the cases and controls. The associations between genotypes and cervical cancer risk were estimated by calculating the odds ratios (ORs) and their 95% confidence intervals (95% CIs) from logistic regression analyses. The adjustment factors for the associations included age, age at menarche, menopausal status and parity. Genotype distributions in the control subjects were checked for Hardy-Weinberg equilibrium by using goodness-of-fit χ2 test. All of the statistical analyses were performed with Statistical Analysis System software (9.1.3; SAS institute, Cary, NC, USA).
Results
Characteristics of study population
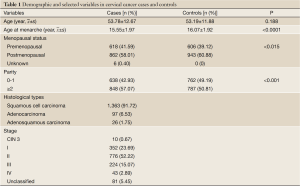
Basic characteristics of the study objects are presented in Table 1. The age was comparable between cases and controls (P>0.05) after frequency-matching. Compared with the control subjects, the cervical cancer cases had an earlier menarche (P<0.05), and showed a lower proportion of menopause and higher parity (P<0.05). Out of the 1,486 cervical cancer cases, 1,363 (91.7%) were squamous cell carcinoma, 97 (6.5%) were adenocarcinoma, 26 (1.8%) were adenosquamous carcinoma. Only 10 (0.7%) patients had cervical intraepithelial neoplasia (CIN) grade 3, 352 (23.7%) were stage I carcinoma, 776 (52.2%) stage II, 224 (15.1%) stage III, 43 (2.9%) stage IV, and 81 (5.5%) unclassified.
Full Table
Genotype distributions of rs3803012, rs1057035 and rs10773771 and cervical cancer risk
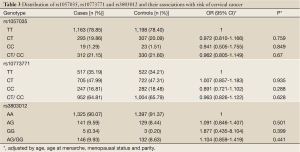
The general characteristics and genotype distributions of RAN rs3803012, DICER rs1057035 and HIWI rs10773771 are shown in Table 2 and Table 3. The genotyping calling rates were 98.8% for rs3803012, 98.9% for rs1057035 and 98.7% for rs10773771, respectively, and the observed genotype frequencies for these three polymorphisms in the controls were all in Hardy-Weinberg equilibrium (P=0.990, 0.513 and 0.245 for rs3803012, rs1057035 and rs10773771 respectively). However, in the multivariate logistic regression models, none of the three SNPs was significantly associated with the risk of cervical cancer. RAN rs3803012 AG/GG vs. AA adjusted OR =1.104, 95% CI: 0.859-1.419; DICER rs1057035 CT/CC vs. TT adjusted OR =0.962, 95% CI: 0.805-1.149; HIWI rs10773771 CT/CC vs. TT adjusted OR =0.963, 95% CI: 0.826-1.122 (Table 3).

Full Table
Full Table
Stratified analyses
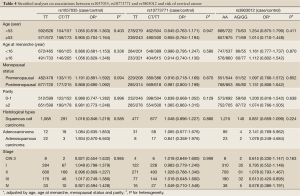
Additionally, we further performed stratification analyses based on age, age at menarche, parity and menopausal status, histological types and tumor stage. As shown in Table 4, the association with rs3803012 was more evident in patients with stage III carcinoma (adjusted OR =0.613; 95% CI: 0.420-0.895). No significant heterogeneity was observed between the stratified subgroups (P>0.100 for heterogeneity tests).
Full Table
Discussion
This study evaluated the relationship of common genetic variants at miRNA biogenesis pathway and the risk of cervical cancer in a Chinese population and found that none of these SNPs was significantly associated with Chinese cervical cancer risk.
DICER plays an important role in the cleavage of pre-miRNAs into their mature form and has previously been implicated in the oncogenic process of several cancers (13-16). To investigate the impact of functional SNPs of DICER gene on tumor development, molecular epidemiological studies were conducted for several cancer types or related diseases, including bladder cancer, lung cancer, renal cell carcinoma, breast cancer and oral premalignant lesions (19-24). However, the results remained inconsistent. Clague et al. reported that rs3742330 in the 3'-UTR of DICER was associated with an increased risk of premalignant oral lesions in Caucasians (OR =2.09, 95% CI: 1.03-4.24, P=0.04) (24). Nevertheless, other studies showed that this SNP in the 3'-UTR of DICER was not associated with risk of cancers such as bladder cancer, lung cancer, renal cell carcinoma and esophageal cancer (19-22). Later, Sung et al. conducted a population-based case control study of 559 breast cancer cases and 567 controls in Korean women and found that rs1057035 in the 3'-UTR of DICER was not associated with risk of breast cancer (OR =0.64, 95% CI: 0.23-1.77, P=0.39) (23). Similarly, our data also did not support an association of cervical cancer risk with the SNP rs1057035 in the 3'-UTR of DICER in Chinese women (OR =0.962, 95% CI: 0.805-1.149).
RAN is a key ingredient for the transportation of pre-miRNAs from nucleus to cytoplasm through the nuclear pore complex in a GTP-dependent manner (25). Overexpression of RAN GTPase has been observed in various other malignancies including stomach, colon, pancreas, lung and ovarian cancer (26-28). The association between RAN polymorphisms and risk of other kind of cancers has been evaluated in several recent studies, but the results were conflicted (19-23). Ye et al. reported that rs14035 in the 3'-UTR of RAN was associated with an increased esophageal cancer risk in Caucasian population (OR =1.99, 95% CI: 1.17-3.38, P=0.011), while other studies reported that this SNP in the 3'-UTR of RAN was not associated with risk of cancers such as bladder cancer, lung cancer and renal cell carcinoma (20-22). In this study, we did not observe the significant association between the rs3803012 in the 3'-UTR of RAN and the risk of cervical cancer.
As a human member of PIWI family, HIWI is suggested to play an important role in stem cell self-renewal, division, RNA silencing and translational regulation (29), and its expression usually upregulated in many types of cancer (17,30,31). Numerous studies have suggested that the rs1106042 (Lys527Arg) in the cording region of HIWI gene was not associated with the risk of cancers such as esophageal cancer, bladder cancer, breast cancer, lung cancer and renal cell carcinoma (19-24). In our study, bioinformatics predict that the SNP rs10773771 (C > T) in the 3'-UTR of HIWI gene could influence the has-miR-1264 binding to HIWI transcript, nevertheless we did not found that this SNP contributed to the risk of cervical cancer.
In the current study, the potential functional SNPs at miRNA biogenesis pathway genes were not significantly associated with the risk of cervical cancer. Although it is difficult to explain the basis for the conflicting results from different studies, the differences of genetic backgrounds and environmental factors in the study populations might contribute to the discrepancy. Another possible explanation is that the discrepancy between the different studies may be that different cell types of cancer have specific etiologies and carcinogenesis pathways. There is increasing evidence that miRNA expression profiles are different according to human cancers (9,11). Therefore, genetic variations in miRNA biosynthesis pathway genes might play different roles in different cell types of cancer.
Several limitations in our study need to be addressed. Firstly, although this is a population based case-control study, the selection bias may not be avoidable and the subjects may not be representative of the general population. Secondly, the polymorphisms conducted in our study, based on their functional considerations, may not give a comprehensive view about genetic variability in miRNA biogenesis pathway genes.
In conclusion, our data did not support an association of cervical risk with genetic variations in the 3'-UTR of RAN, DICER and HIWI genes in a Chinese population. Further tagging and functional SNP-based studies in the whole miRNA biogenesis pathway genes are warranted to confirm and extend the results.
Acknowledgements
Disclosure: The authors declare no conflict of interest.
References
- Arbyn M, Castellsagué X, de Sanjosé S, et al. Worldwide burden of cervical cancer in 2008. Ann Oncol 2011;22:2675-86. [PubMed]
- Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005;55:74-108. [PubMed]
- Wen C. China’s plans to curb cervical cancer. Lancet Oncol 2005;6:139-41. [PubMed]
- Scheurer ME, Tortolero-Luna G, Adler-Storthz K. Human papillomavirus infection: biology, epidemiology, and prevention. Int J Gynecol Cancer 2005;15:727-46. [PubMed]
- Magnusson PK, Lichtenstein P, Gyllensten UB. Heritability of cervical tumours. Int J Cancer 2000;88:698-701. [PubMed]
- Baseman JG, Koutsky LA. The epidemiology of human papillomavirus infections. J Clin Virol 2005;32:S16-24. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Ambros V. The functions of animal microRNAs. Nature 2004;431:350-5. [PubMed]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006;6:857-66. [PubMed]
- Spizzo R, Nicoloso MS, Croce CM, et al. SnapShot: MicroRNAs in Cancer. Cell 2009;137:586-586.e1.
- Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature 2005;435:834-8. [PubMed]
- Kumar MS, Lu J, Mercer KL, et al. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet 2007;39:673-7. [PubMed]
- Chiosea S, Jelezcova E, Chandran U, et al. Up-regulation of dicer, a component of the MicroRNA machinery, in prostate adenocarcinoma. Am J Pathol 2006;169:1812-20. [PubMed]
- Karube Y, Tanaka H, Osada H, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci 2005;96:111-5. [PubMed]
- Faber C, Horst D, Hlubek F, et al. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur J Cancer 2011;47:1414-9. [PubMed]
- Yan M, Huang HY, Wang T, et al. Dysregulated expression of dicer and drosha in breast cancer. Pathol Oncol Res 2012;18:343-8. [PubMed]
- Li S, Meng L, Zhu C, et al. The universal overexpression of a cancer testis antigen hiwi is associated with cancer angiogenesis. Oncol Rep 2010;23:1063-8. [PubMed]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389-402. [PubMed]
- Ye Y, Wang KK, Gu J, et al. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev Res (Phila) 2008;1:460-9. [PubMed]
- Yang H, Dinney CP, Ye Y, et al. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res 2008;68:2530-7. [PubMed]
- Kim JS, Choi YY, Jin G, et al. Association of a common AGO1 variant with lung cancer risk: a two-stage case-control study. Mol Carcinog 2010;49:913-21. [PubMed]
- Horikawa Y, Wood CG, Yang H, et al. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin Cancer Res 2008;14:7956-62. [PubMed]
- Sung H, Lee KM, Choi JY, et al. Common genetic polymorphisms of microRNA biogenesis pathway genes and risk of breast cancer: a case-control study in Korea. Breast Cancer Res Treat 2011;130:939-51. [PubMed]
- Clague J, Lippman SM, Yang H, et al. Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol Carcinog 2010;49:183-9. [PubMed]
- Lund E, Güttinger S, Calado A, et al. Nuclear export of microRNA precursors. Science 2004;303:95-8. [PubMed]
- Xia F, Lee CW, Altieri DC. Tumor cell dependence on Ran-GTP-directed mitosis. Cancer Res 2008;68:1826-33. [PubMed]
- Abe H, Kamai T, Shirataki H, et al. High expression of Ran GTPase is associated with local invasion and metastasis of human clear cell renal cell carcinoma. Int J Cancer 2008;122:2391-7. [PubMed]
- Barrès V, Ouellet V, Lafontaine J, et al. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol Cancer 2010;9:272. [PubMed]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008;9:22-32. [PubMed]
- Grochola LF, Greither T, Taubert H, et al. The stem cell-associated Hiwi gene in human adenocarcinoma of the pancreas: expression and risk of tumour-related death. Br J Cancer 2008;99:1083-8. [PubMed]
- He W, Wang Z, Wang Q, et al. Expression of HIWI in human esophageal squamous cell carcinoma is significantly associated with poorer prognosis. BMC Cancer 2009;9:426. [PubMed]


