Construction of PR domain eukaryotic expression vector and its inhibitory effect on esophageal cancer cells
Introduction
Human retinoblastoma protein-interacting zinc finger gene, RIZ, was first identified from a functional screening for retinoblastoma tumor suppressor binding genes (1,2). RIZ, located on the distal short arm of chromosome 1 (1q36), encodes two different protein products, RIZ1 and RIZ2, which have been identified to be a tumor suppressor and a proto-oncoprotein, respectively (3-6). The expression levels for RIZ1 and RIZ2 are nearly the same among many different human tissues (7). However, an imbalance in the amount of RIZ1 and RIZ2 may be an important cause of cancer development (4). The RIZ1 product is considered as a strong candidate for the tumor suppressor, which commonly undergoes deletions in more than a dozen of different types of human cancers (8). RIZ1 gene expression but not RIZ2 is commonly silenced in all of the human cancers examined including those of breast, liver, colon, and neuroendocrine tissues (9,10).
RIZ1 and RIZ2 have identical amino acid sequences except that RIZ2 lacks the N-terminal PR domain region (PRDM2, about 200 amino acids) (11), which is a member of the PRDM (PRDI-BF1 and RIZ homology domain) family (12). Because of the presence of the PR domain, RIZ is a member of PR/SET gene family. This gene family is known to play an important role in the chromatin-mediated regulation of gene expression during development of cancer, probably through a nuclear protein methylation pathway (13). Therefore, the PR domain is likely responsible for the tumor suppressing activity of RIZ1. Despite RIZ1 mRNA was commonly found absent or at reduced levels in tumor cell lines and tissues while RIZ2 was always found expressed, and in recent studies, few studies of the correlation between RIZ1 and esophageal cancer were reported. Since the PR domain is the only structural difference between the two protein products (RIZ1 and RIZ2) of RIZ gene, we decided to examine whether the PR domain alone possessed any anticancer activity in esophageal cancer cells. In this study, we constructed human PR domain eukaryotic expression vector and transfected human esophageal cancer cells (TE13), and evaluated the anticancer activity of the PR domain against human esophageal cancer TE13 cells.
Materials and methods
Extract mRNA from human esophageal cancer tissue by RT-PCR and reverse-transcribe to cDNA
We started with 100 mg of esophageal cancer tissue cryopreserved at –80 °C, added 1 mL Trizol (Invitrogen corporation, USA), and mixed evenly. The mixture was allowed to stand at room temperature for 5 min, and then centrifuged at 5,000 r/min for 5 min, and the supernatant was transferred into the new Eppendorf (EP) tube. We then added trichlormethane, shook to misce bene, and let the mixture stand for 5 min on ice, and then centrifuged at 12,000 r/min for 15 min at 4 °C. We gently transferred the upper aqueous phase to another new EP tube, added the same volume of isopropanol, then shook the mixture, and let it stand for 5 min at room temperature, centrifuged at 12,000 r/min for 20 min at 4 °C, and removed the supernatant. Next, we added 1 mL 75% ice ethanol prepared with diethylpyrocarbonate (DEPC) into the precipitate, and centrifuged at 7,500 r/min for 5 min at 4 °C, and removed the supernatant. The mixture was allowed to stand for 10 min at room temperature to precipitate fully. Then we dissolved the precipitate in 20 µL DEPC, and determined the concentration of RNA using spectrophotometer at 260 nm. We reverse-transcribed 3 µg RNA using RT-PCR kit under the transcription conditions of: 42 °C 30 min, 85 °C 5 min, cooling to 4 °C, and finally stored at –20 °C.
Amplify PR domain and link to T vector
According to RIZ1 and its sequence published in NCBI, we designed primers to amplify PR domain using Primer6.0 soft (Systhesized by Beijing AuGCT DNA-SYN Biotechnology Co., Ltd.) as follows, forward primer: 5'-gtggctagcATGAATCAGAACACTACTG-3', the 5' end contains Nhe I restriction site and three protective bases; reverse primer: 5'-ttgggatccTCAAGAGGTGAAATCTG-3', the 5' end contains BamH I restriction site and three protective bases, the downstream primer contains a termination codon. KOD plus version 2 [TaKaRa Biotechnology (Dalian) Co., Ltd.] was used with the extracted cDNA as a template. We established a 100 µL PCR amplification system to amplify PR domain by PCR. The PCR parameters were as follow: pre-denaturation at 94 °C for 3 min, 35 cycles of denaturation at 98 °C for 10 s and annealing at 60 °C for 1 min, and elongation at 72 °C in 10 min, at this time, we added Easy Taq enzyme [TianGen Biotech (Beijing) Co., Ltd.] 0.3 µL to add ploy(A) tail to PCR product end, and turned the blunt end into cohesive end. We continued elongation at 72 °C for 30 min, and at last, heat preservation at 4 °C. PCR products were electrophoresed on agarose gel and imaged by the Gel imaging system.
The objective band was recycled by agarose gel DNA recycling purification kit [TianGen Biotech (Beijing) Co., Ltd.]. The recycled PCR products were connected with 1 µL pEASY-T3 cloning vector (Beijing TransGen Biotech Co., Ltd.). We added 30 µL Trans1-T1 phage resistant competent cells into the products connected in preceding step, and mixed them gently. Then we put the mixtures in ice bath for 5 min, followed by a 42 °C water bath, heat shock for 30 s and another ice bath for 5 min. We added 500 µL sterile LB medium into a centrifugal tube, after thorough mixing, we put them in a shaking incubator (constant temperature at 37 °C and 200 r/min) for 1 h for bacteria recovery. The bacteria liquid was centrifuged at 7,000 r/min for 5 min, the supernatant was discarded, and the precipitate was mixed. We added about 100 µL on LB agar medium which contained ampicillin and treated by isopropyl-β-D-1-thiogalactopyranoside (IPTG) and X-gal, smeared the colonies uniformly, and cultured at 37 °C overnight. After 16 h, we picked white colonies by toothpicks and certified by PCR. Then we picked out two colonies whose bands are the brightest, and dropped them into LB liquid medium, then put them on a shaker and cultured at 37 °C overnight. The bacteria liquid was sequenced by Beijing AuGCT DNA-SYN Biotechnology Co., Ltd.
Extract, enzyme cut, link PR domain to pcDNA3.1(+) and transfer into Trans1-T1 phage resistant competent cells
TIANpure Midi Plasmid Kit [TianGen Biotech (Beijing) Co., Ltd.] was used for the PR domain and pcDNA3.1(+) plasmid extraction. We took 43 µL of PR domain plasmid and pcDNA3.1(+) plasmid, respectively, both used BamH I enzyme (1 µL), Nhe I enzyme (1 µL) and NEB buffer 3 (5 µL) [New England Biolabs (Beijing) Ltd.], to set up a 50 µL system, and digested at 37 ºC for 2 h. We recovered the purpose bands, which contained PR domain of more than 600 bp and pcDNA3.1(+) of more than 5,000 bp, by 1.2% agarose gel electrophoresis. We linked PR domain and pcDNA3.1(+) plasmid by use of T4 DNA ligase and 2× rapid ligase DNA buffer. Then we transmitted the product to Trans1-T1 phage resistant competent cells, shook the bacteria at 37 °C overnight. The next day, we picked single bacterial colonies by aseptic toothpicks and performed PCR assay. The bacterial colonies were sequenced.
Culture TE13 cells, extract ultrapure plasmid and transfect in TE13 cells
In order to transfect, we first extracted pcDNA3.1(+)/PR domain plasmid and pcDNA3.1(+) plasmid by TIANpure Midi Plasmid Kit. We recovered esophageal cancer TE13 cells, cultured the cells in incubator at 5% CO2 and 37 °C, and then passaged. TE13 cells, which had been digested by trypsogen, were inoculated in 12-well flat-bottomed plates at a concentration of 1.5×105/mL, cultured to 80% fusion degrees, and prepared for transfection. We set up 6 groups, each group had 4 holes, and the ratio of LipofectamineTM 2000 to plasmid was 2.5 µL to 1 µg. The detailed grouping method is shown in Table 1. After 6 h, we discarded the transfection solution, and added culture medium containing 10% FBS (Gibco, New Zealand). We then cultured the cells for 48 h and collected the results.
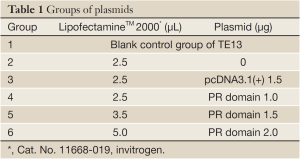
Full Table
The protein expression of pcDNA3.1(+)/PR domain in TE13 by western blotting
After digestion, each group of cells was collected in one centrifuge tube and centrifugated, after removing the supernatant, and twice washed with PBS (0.01 mol/L, pH 7.2-7.3). Each tube of cells was added into 100 µL RIPA lysate with PMSF (Research Organics, INC), allowed to stand for 30 min on ice, and centrifugated at 12,000 r/min for 5 min at 4 °C. The supernatant was removed into a new tube, the protein concentration was determined by BCA protein assay, and the protein sample was preserved at –20 °C. After quantification and aliquot, the samples were degenerated by boiling. Equal amounts of total protein (30 µg/sample) were subjected to SDS-PAGE (12%) followed by electrophoretic transfer to nitrocellulose membranes. To detect membrane protein translocation, each blot was incubated with the first antibody (PRDM2, Abgent, USA), which was used at a dilution of 1:500 for 2 h, and then, with FITC-tagged secondary antibody (l:5,000) for 1 h at room temperature. Then the nitrocellulose membranes were washed 3 times by Tris(hydroxymethyl)aminomethane solution, and subsequently, undertaken to exposure and developing.
Detect apoptosis of TE13 by flow cytometry
Transfection proceeded on a suitable plasmid dosage according to the results of Western blotting. The experimental group was compared with the group of simple TE13 cells and blank plasmid. When growing to 100% fusion degrees, the cells were digested with trypsinase to obtain single cell suspension, and then collected in an EP tube. After centrifugating and discarding the supernatant, the cells were dissolved in 1 mL entire culture solution. Hoechst 33342 was added, and the final concentration was 1 µg/mL, which was incubated at 37 °C for 10 min. The tube was centrifuged at 7,500 r/min for 5 min at 4 °C and the staining solution was discarded. The propidium iodide staining solution was added, and the tube was protected from light at 4 °C for 15 min, followed by filtering with a 400-screen cloth. Flow cytometry was used, and this process was repeated three times.
Statistical analysis
Data are expressed as x̄±s and analyzed by independent-samples t-test with SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). P<0.05 was considered statistically significant.
Results
Construction of PR domain eukaryotic expression vector
We amplified the PR domain by PCR using cDNA as a template. The length of the PCR product was 625 bp, among the total product, the target gene was 600 bp, and the sequence of restriction enzyme recognition site, start codon and termination codon recognition site was 25 bp. More than 600 bp objective band was seen by agarose gel electrophoresis, and this result was consistent with prediction (Figure 1A).
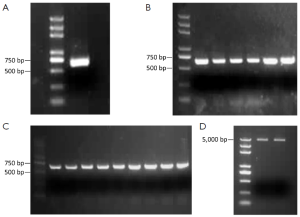
We transferred the PR domain into the Trans1-T1 phage resistant competent cells, shook the bacteria, planked, picked 6 single bacterial colonies, and then performed a PCR assay. More than 600 bp objective band was seen by agarose gel electrophoresis, which was consistent with prediction (Figure 1B).
We transferred the linking product of pcDNA3.1(+) and the PR domain into Trans1-T1 phage resistant competent cells, shook the bacteria, planked, picked 9 single bacterial colonies, and performed a PCR assay. More than 600 bp objective band was seen by agarose gel electrophoresis, which was consistent with prediction (Figure 1C).
We extracted pcDNA3.1/PR domain plasmid, and more than 5,000 bp objective band was seen by agarose gel electrophoresis, which was consistent with prediction (Figure 1D), and the result of sequencing was correct.
Western blotting analysis
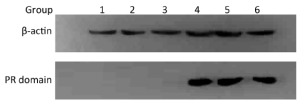
After transfection, the pcDNA3.1(+)/PR domain protein expression in TE13 was detected by Western blotting. The results showed the expression of internal reference β-actin about 42-43 kD in group 1 to 6, which corresponded to Table 1. However, the PR domain whose molecular weight is 28 Da was only expressed in group 4, 5, 6. This result indicated the transfection was successful, and the PR domain can express independently (Figure 2).
Flow cytometry analysis
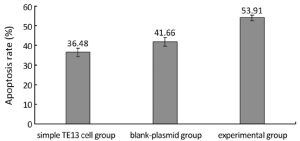
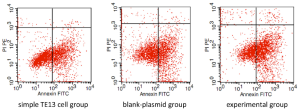
We detected the apoptosis of TE13 by flow cytometry (Figures 3,4). The group of 2.0-µg-PR domain was chosen as an experimental group. The apoptosis rates in the simple TE13 group, the blank-plasmid group and the experimental group were 36.48%±2.11%, 41.66%±2.27% and 53.91%±1.34%, respectively. The apoptosis rate in experimental group was higher than that in the simple TE13 group (P=0.000) and the blank-plasmid group (P=0.001).
Discussion
Esophageal cancers are typically carcinomas which arise from the epithelium, or the surface lining of the esophagus. It is one of the highest incidence malignant tumors in the world, and China is one of the high-incidence areas of all time. A 2008 update by WHO revealed the mortality of esophageal cancer as 5.8/100,000, the eighth highest in the world, but in China, the mortality was 13.4/100,000 and the fourth. Though the therapeutic methods are numerous, treatment effectiveness is far from satisfactory. For the past few years, people gradually began to pay attention to choose the suitable patients and set individualized treatment program from molecular level.
Tumor suppressor genes play an important role in human cancer development. Several tens of tumor suppressor genes have been discovered so far. However, few of these are involved in esophageal cancer. The gene RIZ is a candidate tumor suppressor gene belonging to the PR/SET domain family of chromosomal regulators involved in chromatin-mediated gene activation and silencing. The PR/SET domain family plays an important role in human cancers as evidenced by genetic alterations of several members of this family. The PR family is involved in cancers through an unusual ‘yin-yang’ fashion (14). The PR domain expresses different protein products when it is present or not present, and the imbalance of two products is the early change of gene inactivation. This is also one of main mechanisms of tumorigenesis (15). The two alternative protein products of RIZ are RIZ1, which contains a PR domain at the N-terminal that is involved in tumor suppressor function, and RIZ2, which lacks of this domain and may have a positive role in neoplastic processes (16). The PR domain defines a sub-class of zinc finger gene RIZ1, and was first noted as the PRDI-BF1-RIZ1 homologous region. It contains 200 amino acids. The PR domain may function as a protein binding interface in the regulation of chromatin-mediated gene expression and mediate protein-protein interaction, and plays an important role in the structure stability of chromosome (17).
Since RIZ gene was separated for the first time by Buyse in 1995, the reports about the relationship between RIZ gene and tumors have been endless. Now, some reports have confirmed that RIZ1 gene commonly undergone deletions, rearrangements, or loss of heterozygosity in a broad spectrum of human tumors, including those of breast, liver, colon and neurocrine tissues. Its mechanism may act as a transcription repressor, and an inducer of G2/M cell cycle arrest and/or apoptosis (18). However, the reports about the relationship between RIZ gene and esophageal cancer are seldom. Our preceding experiments confirmed that the expression of mRNA and protein in esophageal cancer was significantly lower than that in normal esophageal tissue. Promoter hypermethylation may play an important role in the epigenetic silencing of RIZ1 gene expression, and 5-Aza-CdR could up-regulate the expression of RIZ1 mRNA in TE13 cell line and inhibit the cell proliferation (19,20). RIZ1 gene can inhibit the occurrence of esophageal cancer, but is very large, so constructing the eukaryotic expression vector is very difficult. The tumor suppressing effect of RIZ1 is concerned with PR domain, therefore, we constructed the PR domain eukaryotic expression vector, and transfected it to TE13 esophageal cancer cells, detected the protein expression of PR domain, and observed its inhibitory effect on esophageal cancer cells.
RIZ1 gene expression in esophageal cancer is low, and the protein expression of PR domain alone should not exist. First of all, we used pcDNA3.1 vector to construct eukaryotic expression vector of PR domain. pcDNA3.1 vector is a kind of high-performance eukaryotic expression vector containing human cytomegalovirus (CMV) strong promoter, which starts the process of gene transcription, in the upstream of insertion sequence, and the downstream is transcription terminate signal. Compared with the virus vector, the pcDNA3.1 has no immunogenicity and potential side effects, and also will not make the virus spread. We linked the PR domain to pcDNA3.1 through enzyme cutting, then transferred it to competent cells to obtain steady amplifiable eukaryotic expression vector of pcDNA3.1/PR domain, and later, transfected it to TE13 cells. Then we got the 28 kD PR domain protein band from TE13 cell lines using Western blotting, and the control group did not get the corresponding band. This fully confirmed that our transfection succeeded and that the PR domain can copy and express stably in TE13 cell lines. Using flow cytometry, we found that the growth and passage of TE13 cells were obviously restrained, and the cell apoptosis was increased. These results showed that the PR domain alone can play an important role in suppressing tumor cells. Our results are consistent with Sun’s experiment (21), which suggested that the PR domain increased the cell death rate in human hepatoma HuH7 cells after transfection. The PR domain alone has anticancer activity. This inhibition of cell proliferation is equivalent to that of RIZ1. Liu’s study indicated that forced expression of RIZ1 results in inhibition of cell proliferation and induction of apoptosis (22).
This experiment successfully constructed PR domain eukaryotic expression vector, and confirmed the inhibition effect on TE13 esophageal cancer cells, which showed the PR domain plays an important role in inhibiting tumor formation. But the experiment also has some shortages. Firstly, the PR domain is only one part of RIZ1, so it cannot represent RIZ1 function completely. Secondly, it is still unclear whether the anticancer activity of the PR domain is due to its methyltransferase activity or the interaction with the PRB motif. Last, this study tested the function of PR domain only in TE13 cell line, but it’s not known whether it would be regulated by other factors in in vivo environment. Consequently, further studies for the PR domain may help to understand the exact mechanism for its anticancer function.
Acknowledgements
This study was supported by National Natural Science Foundation of China (No. 81201945) and Science foundation of Tianjin medical University (No. 2011KY08).
Disclosure: The authors declare no conflict of interest.
References
- Buyse IM, Shao G, Huang S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc Natl Acad Sci USA 1995;92:4467-71. [PubMed]
- Derunes C, Briknarová K, Geng L, et al. Characterization of the PR domain of RIZ1 histone methyltransferase. Biochem Biophys Res Commun 2005;333:925-34. [PubMed]
- Lakshmikuttyamma A, Takahashi N, Pastural E, et al. RIZ1 is potential CML tumor suppressor that is down-regulated during disease progression. J Hematol Oncol 2009;2:28. [PubMed]
- Jiang GL, Huang S. Adenovirus expressing RIZ1 in tumor suppressor gene therapy of microsatellite-unstable colorectal cancers. Cancer Res 2001;61:1796-8. [PubMed]
- Du Y, Carling T, Fang W, et al. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res 2001;61:8094-9. [PubMed]
- Deng Q, Huang S. PRDM5 is silenced in human cancers and has growth suppressive activities. Oncogene 2004;23:4903-10. [PubMed]
- Liu L, Shao G, Steele-Perkins G, et al. The retinoblastoma interacting zinc finger gene RIZ produces a PR domain-lacking product through an internal promoter. J Biol Chem 1997;272:2984-91. [PubMed]
- Huang S. The retinoblastoma protein-interacting zinc finger gene RIZ in 1p36-linked cancers. Front Biosci 1999;4:D528-32. [PubMed]
- Lal G, Padmanabha L, Smith BJ, et al. RIZ1 is epigenetically inactivated by promoter hypermethylation in thyroid carcinoma. Cancer 2006;107:2752-9. [PubMed]
- Jiang GI, Liu L, Buyse IM, et al. Decreased RIZ1 expression but not RIZ2 in hepatoma and suppression of hepatoma tumorigenicity by RIZ1. Int J Cancer 1999;83:541-6. [PubMed]
- Pan KF, Lu YY, Liu WG, et al. Detection of frameshift mutations of RIZ in gastric cancers with microsatellite instability. World J Gastroenterol 2004;10:2719-22. [PubMed]
- Kim KC, Huang S. Histone methyltransferases in tumor suppression. Cancer Biol Ther 2003;2:491-9. [PubMed]
- Gazzerro P, Abbondanza C, D'Arcangelo A, et al. Modulation of RIZ gene expression is associated to estradiol control of MCF-7 breast cancer cell proliferation. Exp Cell Res 2006;312:340-9. [PubMed]
- Jiang GL, Huang S. The yin-yang of PR-domain family genes in tumorigenesis. Histol Histopathol 2000;15:109-17. [PubMed]
- Sun W, Geyer CR, Yang J. Cloning, expression, purification and crystallization of the PR domain of human retinoblastoma protein-binding zinc finger protein 1 (RIZ1). Int J Mol Sci 2008;9:943-50. [PubMed]
- Emterling A, Wallin A, Arbman G, et al. Clinicopathological significance of microsatellite instability and mutated RIZ in colorectal cancer. Ann Oncol 2004;15:242-6. [PubMed]
- Huang S, Shao G, Liu L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J Biol Chem 1998;273:15933-9. [PubMed]
- He L, Yu JX, Liu L, et al. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res 1998;58:4238-44. [PubMed]
- Dong SW, Cui YT, Zhong RR, et al. Decreased expression of retinoblastoma protein-interacting zinc-finger gene 1 in human esophageal squamous cell cancer by DNA methylation. Clin Lab 2012;58:41-51. [PubMed]
- Dong SW, Zhang P, Liu YM, et al. Study on RIZ1 Gene promoter methylation status in human esophageal squamous cell carcinoma. World J Gastroenterol 2012;18:576-82. [PubMed]
- Sun W, Qiao L, Liu Q, et al. Anticancer activity of the PR domain of tumor suppressor RIZ1. Int J Med Sci 2011;8:161-7. [PubMed]
- Liu ZY, Wang JY, Liu HH, et al. Retinoblastoma protein-interacting zinc-finger gene 1 (RIZ1) dysregulation in human malignant meningiomas. Oncogene 2013;32:1216-22. [PubMed]